Volume 22, Number 12—December 2016
Synopsis
Assessing the Epidemic Potential of RNA and DNA Viruses
Figure 4
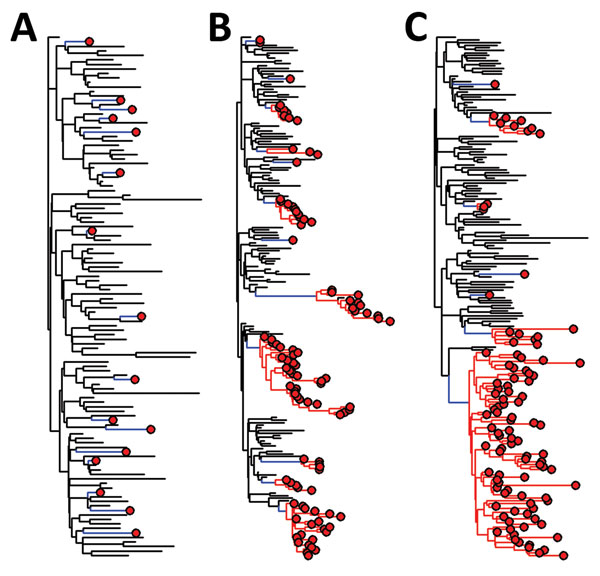
Figure 4. Phylogenetic trees for simulated emerging infectious disease outbreaks caused by RNA and DNA viruses in a mixed population of 1,000 human and 5,000 nonhuman hosts. Trees were constructed by using a standard susceptible–infected–removed model (6). For each of 3 infection scenarios in nonhuman hosts (black lines), rare zoonotic transmission events (blue lines), human-to-human transmission (red lines), and human cases (red circles) are indicated. For the nonhuman population R0 = 2 throughout. Transmissibility within the human populations varies from A) spillover: no human−human transmission (R0 = 0); B) limited human−human transmission with R0 = 1; and C) epidemic spread within humans (R0>1). A maximum of 100 infections are randomly sampled from each population in each simulated outbreak.
References
- Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–9.DOIPubMedGoogle Scholar
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Global trends in emerging infectious diseases. Nature. 2008;451:990–3.DOIPubMedGoogle Scholar
- Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–65.DOIPubMedGoogle Scholar
- King AM, Lefkowitz E, Adams MJ, Carstens EB. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier; 2012.
- Woolhouse ME, Haydon DT, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20:238–44.DOIPubMedGoogle Scholar
- Anderson RM, May RM. Infectious diseases of humans: dynamics and control. New York: Oxford University Press; 1991.
- Hay SI, Battle KE, Pigott DM, Smith DL, Moyes CL, Bhatt S, Global mapping of infectious disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120250.DOIPubMedGoogle Scholar
- Woolhouse ME. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 2002;10(Suppl):S3–7.DOIPubMedGoogle Scholar
- Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–83.DOIPubMedGoogle Scholar
- J Woolhouse ME, Adair K, Brierley L. RNA viruses: a case study of the biology of emerging infectious diseases. Microbiol Spectr. 2013;1:OH-0001–2012.DOIPubMedGoogle Scholar
- Woolhouse ME, Adair K. The diversity of human RNA viruses. Future Virol. 2013;8:159–71 .DOIGoogle Scholar
- Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science. 2006;312:394–7.DOIPubMedGoogle Scholar
- Davies TJ, Pedersen AB. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc Biol Sci. 2008;275:1695–701.DOIPubMedGoogle Scholar
- Cooper N, Nunn CL. 2013 Identifying future zoonotic disease threats. Evol Med Public Health. 2013;1:27–36.DOIPubMedGoogle Scholar
- Khabbaz RF, Heneine W, George JR, Parekh B, Rowe T, Woods T, Brief report: infection of a laboratory worker with simian immunodeficiency virus. N Engl J Med. 1994;330:172–7.DOIPubMedGoogle Scholar
- Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841.DOIPubMedGoogle Scholar
- Chowell G, Hengartner NW, Castillo-Chavez C, Fenimore PW, Hyman JM. The basic reproductive number of Ebola and the effects of public health measures: the cases of Congo and Uganda. J Theor Biol. 2004;229:119–26.DOIPubMedGoogle Scholar
- Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010;8:491–500.DOIPubMedGoogle Scholar
- Fauci AS, Morens DM. Zika virus in the Americas: yet another arbovirus threat. N Engl J Med. 2016;374:601–4.DOIPubMedGoogle Scholar
- Blumberg S, Lloyd-Smith JO. Inference of R(0) and transmission heterogeneity from the size distribution of stuttering chains. PLoS Comput Biol. 2013;9:e1002993.DOIPubMedGoogle Scholar
- Lord CC, Barnard B, Day K, Hargrove JW, McNamara JJ, Paul RE, Aggregation and distribution of strains in microparasites. Philos Trans R Soc Lond B Biol Sci. 1999;354:799–807.DOIPubMedGoogle Scholar
- Yates A, Antia R, Regoes RR. How do pathogen evolution and host heterogeneity interact in disease emergence? Proc Biol Sci. 2006;273:3075–83.DOIPubMedGoogle Scholar
- Jansen VA, Stollenwerk N, Jensen HJ, Ramsay ME, Edmunds WJ, Rhodes CJ. Measles outbreaks in a population with declining vaccine uptake. Science. 2003;301:804.DOIPubMedGoogle Scholar
- Woolhouse ME, Gaunt E. Ecological origins of novel human pathogens. In: Relman DA, Hamburg MA, Choffnes ER, Mack A, editors. Microbial evolution and co-adaptation, Washington (DC): National Academies Press; 2009. p. 208–29.
- Breban R, Riou J, Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382:694–9.DOIPubMedGoogle Scholar
- Fine PE, Jezek Z, Grab B, Dixon H. The transmission potential of monkeypox virus in human populations. Int J Epidemiol. 1988;17:643–50.DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Outbreaks chronology: Ebola virus disease [cited 2015 Feb 1]. http://www.cdc.gov/vhf/ebola/outbreaks/history/chronology.html#modalIdString_outbreaks
- Woolhouse M, Antia R. Emergence of new infectious diseases. In: Stearns SC, Koella JK, editors. Evolution in health and disease. 2nd ed. Oxford (UK): Oxford University Press; 2008. p. 215–28.
- Domingo E, Martínez-Salas E, Sobrino F, de la Torre JC, Portela A, Ortín J, The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene. 1985;40:1–8.DOIPubMedGoogle Scholar
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–41.DOIPubMedGoogle Scholar
- Arinaminpathy N, McLean AR. Evolution and emergence of novel human infections. Proc Biol Sci. 2009;276:3937–43.DOIPubMedGoogle Scholar
- Cotten M, Watson SJ, Zumla AI, Makhdoom HQ, Palser AL, Ong SH, Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. 2014;5:e01062–13.DOIPubMedGoogle Scholar
- Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–72.DOIPubMedGoogle Scholar
- Jombart T, Cori A, Didelot X, Cauchemez S, Fraser C, Ferguson N. Bayesian reconstruction of disease outbreaks by combining epidemiologic and genomic data. PLoS Comput Biol. 2014;10:e1003457.DOIPubMedGoogle Scholar
- Volz EM, Kosakovsky Pond SL, Ward MJ, Leigh Brown AJ, Frost SD. Phylodynamics of infectious disease epidemics. Genetics. 2009;183:1421–30.DOIPubMedGoogle Scholar
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–97.DOIPubMedGoogle Scholar
- Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–4.DOIPubMedGoogle Scholar
- World Health Organization. Blueprint for R&D preparedness and response to public health emergencies due to highly infectious pathogens, 2015 [cited 2016 Aug 21]. http://www.who.int/csr/research-and-development/meeting-report-prioritization.pdf?ua=1
- Belshaw R, Gardner A, Rambaut A, Pybus OG. Pacing a small cage: mutation and RNA viruses. Trends Ecol Evol. 2008;23:188–93.DOIPubMedGoogle Scholar
- Woolhouse ME, Rambaut A, Kellam P. Lessons from Ebola: Improving infectious disease surveillance to inform outbreak management. Sci Transl Med. 2015;7:307rv5.DOIPubMedGoogle Scholar