Volume 22, Number 12—December 2016
Dispatch
Highly Divergent Dengue Virus Type 2 in Traveler Returning from Borneo to Australia
Figure 1
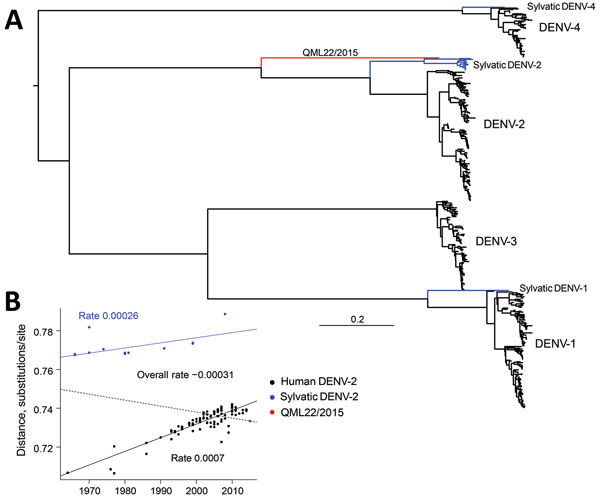
Figure 1. A) Maximum-likelihood phylogenetic tree of 500 complete genome sequences of DENV-1–DENV-4 (alignment length of 10,185 nt), including QML22/2015, estimated by using the generalized time-reversible invariable sites gamma model of nucleotide substitution in PhyML (13) and nearest-neighbor interchange plus subtree pruning, and regrafting branch-swapping. The tree is midpoint rooted for clarity, and sequences are color coded according to their putative transmission cycle (human, black; sylvatic, blue); red indicates the highly divergent QML22/2015 lineage isolated in this study. B) Regression of root-to-tip genetic distances of 119 representative human and sylvatic complete genome sequences of DENV-2 (alignment length 10,173 nt) against time (year) of sampling. The input phylogenetic tree was estimated by using the same maximum-likelihood procedure. Three regression lines and slopes are shown; slopes indicate an estimate of the virus nucleotide substitution rate (substitutions/site/year). Blue line indicates rate for entire DENV-2 data set; dashed line indicates rate for sylvatic DENV-2 sequences; black line indicates rate for human DENV-2 sequences. There is a marked difference between sylvatic and human rates. DENV, dengue virus.
References
- Rudnick A, Marchette NJ, Garcia R. Possible jungle dengue—recent studies and hypotheses. Jpn J Med Sci Biol. 1967;20(Suppl):69–74.PubMedGoogle Scholar
- Kimura R, Hotta S. On the inoculation of dengue virus into mice. Nippon Igaku. 1944;3379:629–33.
- Sabin AB. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1:30–50.PubMedGoogle Scholar
- Hammon WM, Rudnick A, Sather GE. Viruses associated with epidemic hemorrhagic fevers of the Philippines and Thailand. Science. 1960;131:1102–3.DOIPubMedGoogle Scholar
- Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74:3227–34.DOIPubMedGoogle Scholar
- Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol. 2011;9:532–41.DOIPubMedGoogle Scholar
- Pyke AT, Moore PR, Taylor CT, Hall-Mendelin S, Cameron JN, Hewitson GR, et al. Highly divergent dengue virus type 1 genotype sets a new distance record. Sci Rep. 2016;6:22356.DOIPubMedGoogle Scholar
- Roehrig JT, Mathews JH, Trent DW. Identification of epitopes on the E glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology. 1983;128:118–26.DOIPubMedGoogle Scholar
- Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–6.PubMedGoogle Scholar
- Jianmin Z, Linn ML, Bulich R, Gentry MK, Aaskov JG. Analysis of functional epitopes on the dengue 2 envelope (E) protein using monoclonal IgM antibodies. Arch Virol. 1995;140:899–913.DOIPubMedGoogle Scholar
- Sambrook J, Rusell DW, editors. Molecular cloning: a laboratory manual. Cold Spring Harbor (NY): Cold Spring Harbor Press; 2001.
- Biebricher CK, Luce R. Sequence analysis of RNA species synthesized by Q beta replicase without template. Biochemistry. 1993;32:4848–54.DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.DOIPubMedGoogle Scholar
- Popescu AA, Huber KT, Paradis E. ape 3.0: New tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics. 2012;28:1536–7.DOIPubMedGoogle Scholar
- Vasilakis N, Cardosa J, Diallo M, Sall AA, Holmes EC, Hanley KA, et al. Sylvatic dengue viruses share the pathogenic potential of urban/endemic dengue viruses. J Virol. 2010;84:3726–7, author reply 3727–8.DOIPubMedGoogle Scholar