Volume 22, Number 4—April 2016
Letter
Enterovirus A71 Genogroups C and E in Children with Acute Flaccid Paralysis, West Africa
Figure
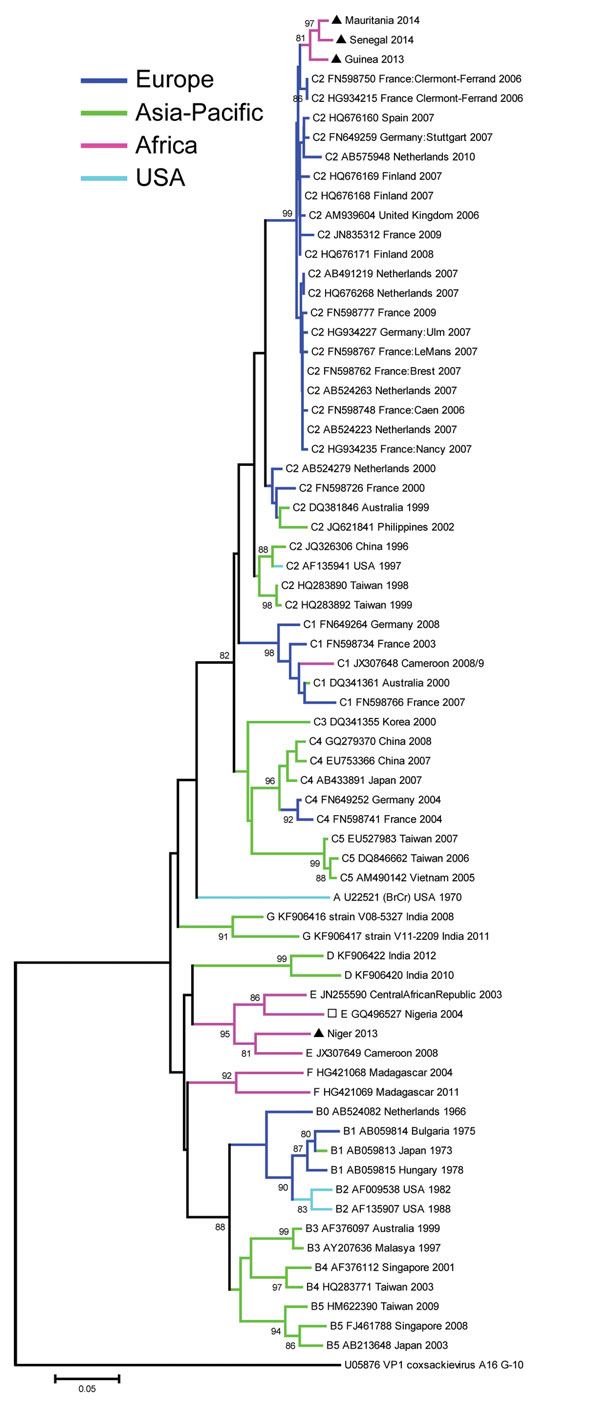
Figure. Phylogenetic tree created with the complete VP1 nucleotide sequences (891 bp in length) of enterovirus A71 isolated from 4 patients with acute flaccid paralysis in West Africa, the most similar nucleotide sequences identified by a search in GenBank by using BLAST (http://www.ncbi.nlm.nih.gov/), and a representative global set of enterovirus A71 sequences belonging to different genogroups and subgenogroups. The coxsackievirus A16 prototype G-10 sequence was introduced as the outgroup. The tree was inferred with a neighbor-joining method that used MEGA5 software (http://www.megasoftware.net/). Distances were computed by using the Kimura 2–parameter model. The robustness of the nodes was tested by using 1,000 bootstrap replications. Bootstrap support values >80 are shown in nodes. The 4 black triangles indicate the 4 strains from this study. The open square represents a partial sequence. Scale bar represents nucleotide substitutions per site. Abbreviations of virus names indicate genogroups or subgenogroups/GenBank accession number/origin/year of isolation, respectively.