Volume 22, Number 6—June 2016
Research
Use of Population Genetics to Assess the Ecology, Evolution, and Population Structure of Coccidioides
Figure 1
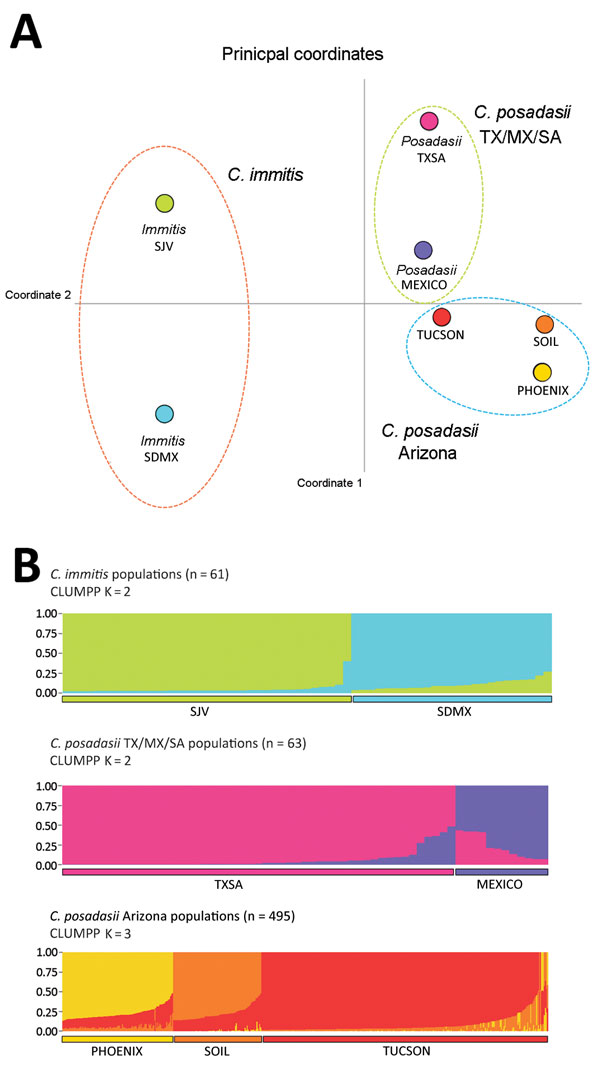
Figure 1. Results of principal coordinate analysis and STRUCTURE analyses of Coccidioides spp. populations. A) Principal coordinate analysis using Nei’s unbiased genetic distance estimates supports 3 main groupings: C. immitis, C. posadasii TX/SA/MX, and C. posadasii Arizona (see also Technical Appendix 3 Figure 2). The greatest separation occurs between species and is reflected in principal coordinate 1 (93.92% of variance). Color-coding for populations: lime green, San Joaquin Valley (SJV); aqua, San Diego/Mexico (SDMX); pink, Texas/South America (TXSA); purple, Mexico (MEXICO); red, Tucson (TUCSON); yellow, Phoenix/Yuma (PHOENIX); orange, soil (AZSOIL). B) STRUCTURE analysis. Microsatellite matrices were analyzed with STRUCTURE 2.3.4 to determine population structure within Coccidioides populations (30). The running length of burn-in period was 100,000 repetitions with 1 million Markov chain Monte Carlo repetitions. Default settings in STRUCTURE 2.3.4 were as follows: the admixture model was used to infer α along with the previous sampling location information model (LOCPRIOR) (30). We used CLUMPP, a cluster matching and permutation program (https://web.stanford.edu/group/rosenberglab/clumpp.html), to define populations within the STRUCTURE algorithm. K is the number of significant populations in each main group. A consensual STRUCTURE plot was generated from the admixture values by using the Clustering Markov Packager Across K (CLUMPAK) server, and final plots were built with STRUCTURE PLOT (32,33).
References
- Fisher MC, Koenig GL, White TJ, Taylor JW. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia. 2002;94:73–84. DOIPubMedGoogle Scholar
- Cairns L, Blythe D, Kao A, Pappagianis D, Kaufman L, Kobayashi J, Outbreak of coccidioidomycosis in Washington state residents returning from Mexico. Clin Infect Dis. 2000;30:61–4. DOIPubMedGoogle Scholar
- Nguyen C, Barker BM, Hoover S, Nix DE, Ampel NM, Frelinger JA, Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of coccidioidomycosis. Clin Microbiol Rev. 2013;26:505–25. DOIPubMedGoogle Scholar
- Lewis ER, Bowers JR, Barker BM. Dust devil: the life and times of the fungus that causes Valley fever. PLoS Pathog. 2015;11:e1004762. DOIPubMedGoogle Scholar
- Huppert M, Sun SH, Harrison JL. Morphogenesis throughout saprobic and parasitic cycles of Coccidioides immitis. Mycopathologia. 1982;78:107–22. DOIPubMedGoogle Scholar
- Chiller TM, Galgiani JN, Stevens DA. Coccidioidomycosis. [viii. ]. Infect Dis Clin North Am. 2003;17:41–57. DOIPubMedGoogle Scholar
- Fisher MC, Koenig GL, White TJ, San-Blas G, Negroni R, Alvarez IG, Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc Natl Acad Sci U S A. 2001;98:4558–62. DOIPubMedGoogle Scholar
- Marsden-Haug N, Hill H, Litvintseva AP, Engelthaler DM, Driebe EM, Roe CC, Coccidioides immitis identified in soil outside of its known range—Washington, 2013. MMWR Morb Mortal Wkly Rep. 2014;63:450 .PubMedGoogle Scholar
- Litvintseva AP, Marsden-Haug N, Hurst S, Hill H, Gade L, Driebe EM, Valley fever: finding new places for an old disease: Coccidioides immitis found in Washington State soil associated with recent human infection. Clin Infect Dis. 2015;60:e1–3. DOIPubMedGoogle Scholar
- Johnson SM, Carlson EL, Fisher FS, Pappagianis D. Demonstration of Coccidioides immitis and Coccidioides posadasii DNA in soil samples collected from Dinosaur National Monument, Utah. Med Mycol. 2014;52:610–7 and. DOIPubMedGoogle Scholar
- Canteros CE, Vélez HA, Toranzo AI, Suárez-Alvarez R, Tobón OÁ, Jimenez AMP, Molecular identification of Coccidioides immitis in formalin-fixed, paraffin-embedded (FFPE) tissues from a Colombian patient. Med Mycol. 2015;53:520–7. DOIPubMedGoogle Scholar
- Whiston E, Taylor JW. Genomics in Coccidioides: insights into evolution, ecology, and pathogenesis. Med Mycol. 2014;52:149–55. DOIPubMedGoogle Scholar
- Duarte-Escalante E, Zúñiga G, Frías-De-León MG, Canteros C, Castañón-Olivares LR, Reyes-Montes MR. AFLP analysis reveals high genetic diversity but low population structure in Coccidioides posadasii isolates from Mexico and Argentina. BMC Infect Dis. 2013;13:411. DOIPubMedGoogle Scholar
- Brilhante RS, de Lima RA, Ribeiro JF, de Camargo ZP, Castelo-Branco DS, Grangeiro TB, Genetic diversity of Coccidioides posadasii from Brazil. Med Mycol. 2013;51:432–7. DOIPubMedGoogle Scholar
- Campins H. Coccidioidomycosis in South America. A review of its epidemiology and geographic distribution. Mycopathol Mycol Appl. 1970;41:25–34. DOIPubMedGoogle Scholar
- Neafsey DE, Barker BM, Sharpton TJ, Stajich JE, Park DJ, Whiston E, Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 2010;20:938–46. DOIPubMedGoogle Scholar
- Fisher MC, Koenig G, White TJ, Taylor JW. A test for concordance between the multilocus genealogies of genes and microsatellites in the pathogenic fungus Coccidioides immitis. Mol Biol Evol. 2000;17:1164–74. DOIPubMedGoogle Scholar
- Fisher MC, White TJ, Taylor JW. Primers for genotyping single nucleotide polymorphisms and microsatellites in the pathogenic fungus Coccidioides immitis. Mol Ecol. 1999;8:1082–4. DOIPubMedGoogle Scholar
- Lim S, Notley-McRobb L, Lim M, Carter DA. A comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genet Biol. 2004;41:1025–36. DOIPubMedGoogle Scholar
- Fisher MC. DE Hoog S, Akom NV. A highly discriminatory multilocus microsatellite typing (MLMT) system for Penicillium marneffei. Mol Ecol Notes. 2004;4:515–8. DOIPubMedGoogle Scholar
- Fisher MC, Aanensen D, de Hoog S, Vanittanakom N. Multilocus microsatellite typing system for Penicillium marneffei reveals spatially structured populations. J Clin Microbiol. 2004;42:5065–9. DOIPubMedGoogle Scholar
- Taylor ML, Hernández-García L, Estrada-Bárcenas D, Salas-Lizana R, Zancopé-Oliveira RM, García de la Cruz S, Genetic diversity of Histoplasma capsulatum isolated from infected bats randomly captured in Mexico, Brazil, and Argentina, using the polymorphism of (GA)(n) microsatellite and its flanking regions. Fungal Biol. 2012;116:308–17.
- Matute DR, Sepulveda VE, Quesada LM, Goldman GH, Taylor JW, Restrepo A, Microsatellite analysis of three phylogenetic species of Paracoccidioides brasiliensis. J Clin Microbiol. 2006;44:2153–7. DOIPubMedGoogle Scholar
- Barker BM, Jewell KA, Kroken S, Orbach MJ. The population biology of coccidioides: epidemiologic implications for disease outbreaks. Ann N Y Acad Sci. 2007;1111:147–63. DOIPubMedGoogle Scholar
- Jewell K, Cheshier R, Cage GD. Genetic diversity among clinical Coccidioides spp. isolates in Arizona. Med Mycol. 2008;46:449–55. DOIPubMedGoogle Scholar
- Hector RF, Rutherford GW, Tsang CA, Erhart LM, McCotter O, Anderson SM, The public health impact of coccidioidomycosis in Arizona and California. Int J Environ Res Public Health. 2011;8:1150–73. DOIPubMedGoogle Scholar
- Fisher MC, Koenig GL, White TJ, Taylor JW. Pathogenic clones versus environmentally driven population increase: analysis of an epidemic of the human fungal pathogen Coccidioides immitis. J Clin Microbiol. 2000;38:807–13 .PubMedGoogle Scholar
- Barker BM, Tabor JA, Shubitz LF, Perrill R, Orbach MJ. Detection and phylogenetic analysis of Coccidioides posadasii in Arizona soil samples. Fungal Ecol. 2012;5:163–76. DOIGoogle Scholar
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–9. DOIPubMedGoogle Scholar
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–32.
- Earl D, vonHoldt B. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–61.
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015;15:1179–91.
- Ramasamy RK, Ramasamy S, Bindroo BB, Naik VG. STRUCTURE PLOT: a program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus. 2014;3:431.
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. DOIPubMedGoogle Scholar
- Baptista-Rosas RC, Hinojosa A, Riquelme M. Ecological niche modeling of Coccidioides spp. in western North American deserts. Ann N Y Acad Sci. 2007;1111:35–46. DOIPubMedGoogle Scholar
- Fisher FS, Bultman MW, Johnson SM, Pappagianis D, Zaborsky E. Coccidioides niches and habitat parameters in the southwestern United States: a matter of scale. Ann N Y Acad Sci. 2007;1111:47–72. DOIPubMedGoogle Scholar
- Litvintseva AP, Brandt ME, Mody RK, Lockhart SR. Investigating fungal outbreaks in the 21st century. PLoS Pathog. 2015;11:e1004804. DOIPubMedGoogle Scholar
- Muller LA, Lucas JE, Georgianna DR, McCusker JH. Genome-wide association analysis of clinical vs. nonclinical origin provides insights into Saccharomyces cerevisiae pathogenesis. Mol Ecol. 2011;20:4085–97. DOIPubMedGoogle Scholar
- Thompson GR III, Stevens DA, Clemons KV, Fierer J, Johnson RH, Sykes J, Call for a California coccidioidomycosis consortium to face the top ten challenges posed by a recalcitrant regional disease. Mycopathologia. 2015;179:1–9. DOIPubMedGoogle Scholar