Volume 23, Number 10—October 2017
Dispatch
Diagnosis of Fatal Human Case of St. Louis Encephalitis Virus Infection by Metagenomic Sequencing, California, 2016
Abstract
We used unbiased metagenomic next-generation sequencing to diagnose a fatal case of meningoencephalitis caused by St. Louis encephalitis virus in a patient from California in September 2016. This case is associated with the recent 2015–2016 reemergence of this virus in the southwestern United States.
St. Louis encephalitis virus (SLEV), in the Flaviviridae family, is an infectious RNA virus transmitted by Culex spp. mosquitoes (1,2). Clinical manifestations range from mild febrile illness to fatal neurologic disease. According to recent reports (3,4), SLEV reemerged in the summer of 2015 in California and Arizona, USA, after a documented 11-year absence of activity in California.
In June 2016, we launched a multi-institutional clinical study titled Precision Diagnosis of Acute Infectious Diseases (PDAID). This 1-year study aimed to enroll 300 patients to evaluate the clinical utility of a metagenomic next-generation sequencing (mNGS) assay for diagnosing infectious causes of meningitis and encephalitis from patient cerebrospinal fluid (CSF) samples (5,6). The mNGS assay uses an unbiased sequencing approach to comprehensively identify pathogens (viruses, bacteria, fungi, and parasites) in clinical samples (7). We report a fatal human case of SLEV infection diagnosed by mNGS in a PDAID study patient from California.
The case-patient was a 68-year-old man with a history of coronary artery disease, hypertension, and mantle cell lymphoma treated with 4 cycles of chemotherapy and granulocyte colony stimulating factor. He sought medical care at the end of August 2016 having had 2 days of fever (up to 39.4°C), chills, lethargy, and confusion. He had fallen twice because of dizziness and reported shortness of breath, cough, and new-onset urinary incontinence.
The patient was a retired oil-field worker living with his wife in Bakersfield, Kern County, California. He owned 1 dog and had frequent contact with his 10 grandchildren. His travel history was notable for a trip to “the mountains” in late April 2016 (Payson, Arizona, elevation 1,500 m).
The patient was admitted to the hospital in early September 2016. An initial workup, including magnetic resonance imaging of the brain, was unrevealing. Empirical therapy with vancomycin, meropenem, and levofloxacin was started after lung imaging revealed inflammatory pneumonitis. On hospitalization day 3, the patient became acutely hypoxic with worsening altered mental status (AMS), and he was intubated and transferred to the intensive care unit. A lumbar puncture revealed CSF pleocytosis (18 leukocytes/mm3; 35% monocytes, 33% lymphocytes, and 32% neutrophils); glucose and protein were within reference ranges. Empiric antibiotic therapy was continued, and acyclovir and antifungal therapy were added to his regimen. Repeat lumbar puncture performed on hospital day 9 showed persistent pleocytosis. All microbiologic test results for CSF, blood, and bronchoalveolar lavage were negative (Table 1), as was a workup for noninfectious causes (Technical Appendix).
After enrolling the patient in the PDAID study, we analyzed CSF from hospitalization day 9 by mNGS testing at University of California, San Francisco (Technical Appendix) (8). RNA and DNA sequencing libraries from CSF yielded 8,056,471 and 9,083,963 sequence reads, respectively. In the RNA library, 236,615 (2.9%) of the reads were identified as SLEV by using the SURPI+ (sequence-based ultra-rapid pathogen identification) computational pipeline (7), with recovery of 99.4% of the predicted 10,936-bp virus genome. Subsequent mNGS testing of the patient’s CSF from hospitalization day 3 also was positive for SLEV.
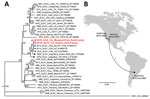
The patient’s SLEV genome sequence was >99% identical with previously sequenced 2014–2015 SLEV isolates from mosquitoes in California and Arizona (4). Phylogenetic analysis placed the patient’s strain in a cluster containing these isolates and viruses sequenced from mosquitoes in Argentina in 1978 and 2005 (9) (Figure). The patient’s SLEV was genetically distinct from the 2003 Imperial Valley strain that had been circulating in California before an 11-year absence (12), suggesting that he was infected by the 2015–2016 reemergent genotype currently circulating in the southwestern United States (3,4). Furthermore, the patient’s SLEV genome was closely related to a strain sequenced from a mosquito pool collected in June 2016 from Kern County (Figure, panel A), with 99.9% pairwise nucleotide identity and only 5 single-nucleotide variants across the genome.
After extensive discussion with his wife regarding the patient’s SLEV diagnosis and poor prognosis, the patient was transitioned to comfort care on hospitalization day 23 and died the following day. Autopsy revealed residual mantle cell lymphoma and bronchopneumonia consistent with infection or chemical pneumonitis from aspiration. The diagnosis of SLEV meningoencephalitis was subsequently confirmed by positive reverse transcription PCR and virus culture testing from multiple laboratories (Table 2). However, follow-up testing for SLEV from the patient’s CSF and serum was negative.
We present a case of SLEV infection in an elderly immunocompromised patient hospitalized with fever and AMS and who experienced critical respiratory failure. Most SLEV infections are asymptomatic; when infections are symptomatic, clinical features include fever, lethargy, and confusion (1), with potential complications including sepsis, gastrointestinal hemorrhage, pulmonary embolism, and bronchopneumonia. In hindsight, SLEV infection is consistent with the patient’s clinical presentation, with pneumonitis and respiratory decompensation possibly resulting from aspiration during the patient’s AMS from viral meningoencephalitis. Deaths from SLEV infection during the first 2 weeks are generally from encephalitis, whereas later deaths are more often caused by complications of hospitalization, such as this patient’s bronchopneumonia.
Routine diagnosis of SLEV is challenging because serologic testing is only performed by specialized reference laboratories, the period of viremia is brief, and molecular testing by reverse transcription PCR is not widely used. Clinicians in California might fail to consider SLEV when examining a patient with nonspecific febrile illness, especially given the lack of virus or disease activity in the state during 2004–2015. Antibody testing can be complicated by the absence of seroreactivity in elderly and immunocompromised patients, as observed in the case of this patient (Table 1), as well as potential cross-reactivity with other flavivirus infections, such as dengue, Zika virus, and West Nile virus (3).
The identification of SLEV infection in CSF by using a panpathogen metagenomic sequencing assay is another demonstration of the power of an unbiased approach to infectious disease testing (5–7), although challenges remain with respect to test availability, interpretation, and validation (8). No antiviral therapy for SLEV has been proven to be efficacious, although interferon-α has been tried (13). With a laboratory sample-to-reporting time of 4 days, earlier sample submission might have spared our patient from the side effects of antimicrobial drug therapy, costly laboratory testing, and invasive procedures. Importantly, the family obtained reassurance and closure from communication of an established diagnosis.
During summer 2016, SLEV was reported in mosquitoes from 7 counties in California, including Kern County, where the patient resided (4). According to his wife, the patient often sat outdoors during the few weeks before hospitalization, although she did not recall his reporting any mosquito bites. Nevertheless, we believe he most likely contracted SLEV in California, because his history of travel to Arizona 5 months prior was not consistent with the incubation period of the disease (4–21 days); mosquitoes are less prevalent at the higher altitudes of Payson, Arizona; and the patient’s SLEV sequence was most closely related to a strain from a June 2016 Kern County mosquito pool. Given the reemergence of SLEV in the southwestern United States, clinicians from affected areas will need to maintain a high index of suspicion for this disease, particularly during local community outbreaks or high SLEV activity detected through mosquito surveillance efforts.
Dr. Chiu is an associate professor at the University of California, San Francisco, who heads a translational research laboratory focused on clinical metagenomic assay development for infectious diseases and characterization of emerging outbreak viruses.
Acknowledgments
We thank staff at the California Department of Health for their assistance in this study, including Kristina Hsieh and Artem Muradyan for validating and performing the SLEV reverse transcription PCR assay and Chris Preas and Peter Patiris for performing IgM and plaque-reduction neutralization serologic tests for SLEV and West Nile virus. We also thank the Centers for Disease Control and Prevention for confirmatory testing of the patient sample for SLEV.
The study was supported by the California Initiative to Advance Precision Medicine (C.Y.C., H.A.S., M.R.W., S.M.); an award from Abbott Laboratories, Inc. (C.Y.C.); and philanthropic grants from the Schwab Foundation (C.Y.C.) and the Sandler and William K. Bowes Foundations (C.Y.C., H.A.S., M.R.W., S.M.).
C.Y.C. is the director of the UCSF–Abbott Viral Diagnostics and Discovery Center and receives research support from Abbott Laboratories, Inc. C.Y.C., S.N.N., and S.M. are co-inventors of the sequence-based ultra-rapid pathogen identification computational pipeline (known as SURPI+) and associated algorithms; a patent pending for SURPI+ has been filed by the University of California, San Francisco.
References
- Salimi H, Cain MD, Klein RS. Encephalitic arboviruses: emergence, clinical presentation, and neuropathogenesis. Neurotherapeutics. 2016;13:514–34. DOIPubMedGoogle Scholar
- CDC. Saint Louis encephalitis [cited 2016 Dec 8]. http://www.cdc.gov/sle
- Venkat H, Krow-Lucal E, Hennessey M, Jones J, Adams L, Fischer M, et al. Concurrent outbreaks of St. Louis encephalitis virus and West Nile virus disease—Arizona, 2015. MMWR Morb Mortal Wkly Rep. 2015;64:1349–50.PubMedGoogle Scholar
- White GS, Symmes K, Sun P, Fang Y, Garcia S, Steiner C, et al. Reemergence of St. Louis encephalitis virus, California, 2015. Emerg Infect Dis. 2016;22:2185–8. DOIPubMedGoogle Scholar
- Mongkolrattanothai K, Naccache SN, Bender JM, Samayoa E, Pham E, Yu G, et al. Neurobrucellosis: unexpected answer from metagenomic next-generation sequencing. J Pediatric Infect Dis Soc. 2017;•••:piw066; Epub ahead of print. DOIPubMedGoogle Scholar
- Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–17. DOIPubMedGoogle Scholar
- Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–92. DOIPubMedGoogle Scholar
- Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists. Microbiology Resource Committee of the College of American Pathologists. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–86. DOIPubMedGoogle Scholar
- Spinsanti LI, Díaz LA, Glatstein N, Arselán S, Morales MA, Farías AA, et al. Human outbreak of St. Louis encephalitis detected in Argentina, 2005. J Clin Virol. 2008;42:27–33. DOIPubMedGoogle Scholar
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. DOIPubMedGoogle Scholar
- Reisen WK, Lothrop HD, Wheeler SS, Kennsington M, Gutierrez A, Fang Y, et al. Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003-2006. J Med Entomol. 2008;45:494–508.PubMedGoogle Scholar
- Rahal JJ, Anderson J, Rosenberg C, Reagan T, Thompson LL. Effect of interferon-alpha2b therapy on St. Louis viral meningoencephalitis: clinical and laboratory results of a pilot study. J Infect Dis. 2004;190:1084–7. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 23, Number 10—October 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Charles Y. Chiu, University of California, San Francisco, Laboratory Medicine, 185 Berry St, Box 0134, UCSF China Basin, San Francisco, CA 94143-0134, USA
Top