Volume 23, Number 10—October 2017
Dispatch
Enterovirus D68–Associated Acute Flaccid Myelitis in Immunocompromised Woman, Italy
Figure 2
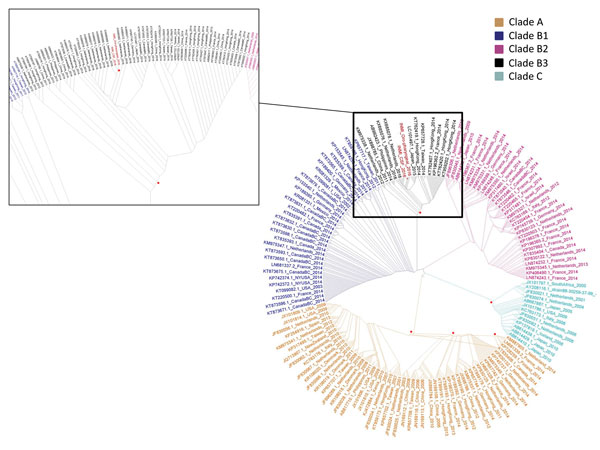
Figure 2. Phylogenetic tree of partial viral protein 1 sequences of 279 nt (nt positions 2581–2859, reference sequence GenBank accession no.AY426531.1), sequences from cerebrospinal fluid (accession no. MF061604) and oropharyngeal swab sample (accession no. MF061605) from a woman who died of fatal neurologic disease associated with enterovirus-D68 infection (indicated in red) in the context of 918 enterovirus-D68 global sequences retrieved from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). The main figure shows the whole maximum-likelihood tree (1,000 bootstrap replicas), generated by using the HKY+GI (Hasegawa-Kishino-Yano + gamma distribution invariant sites) model; the inset shows an enlargement of subclade B3 containing the sequences from the patient reported here and patients involved in the 2016 epidemic in the Netherlands: the closest sequence is KX685068.1_Netherlands_2016 (98% identity, distance: 180 substitutions/104 positions). Red dots indicate nodes with bootstrap value >70.