Volume 23, Number 2—February 2017
Research Letter
Hepatitis E Virus Infection in Solid Organ Transplant Recipients, France
Figure
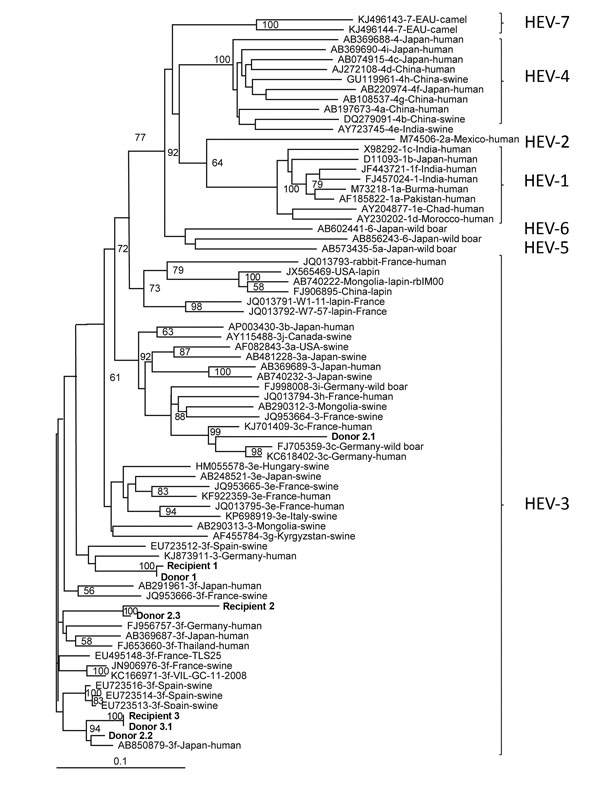
Figure. Phylogenetic tree of hepatitis E virus (HEV) isolates from 3 HEV-positive blood donors and 3 solid organ transplant recipients (shown in bold), France, compared with reference isolates. The tree was constructed by using partial open reading frame 2 sequences (348 nt). HEV genotypes are indicated at right. A confirmed case of transfusion-transmitted HEV infection requires evidence of infection in the recipient and donor and that the nucleotide sequences of these isolates be identical. The isolates from France were deposited in GenBank under accession nos. KX452928–KX452935; accession numbers, sources, and location of isolation for other isolates are indicated. Scale bar indicates nucleotide substitutions per site.
Page created: January 18, 2017
Page updated: January 18, 2017
Page reviewed: January 18, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.