Volume 23, Number 6—June 2017
Research
Genomic Analysis of Salmonella enterica Serovar Typhimurium DT160 Associated with a 14-Year Outbreak, New Zealand, 1998–2012
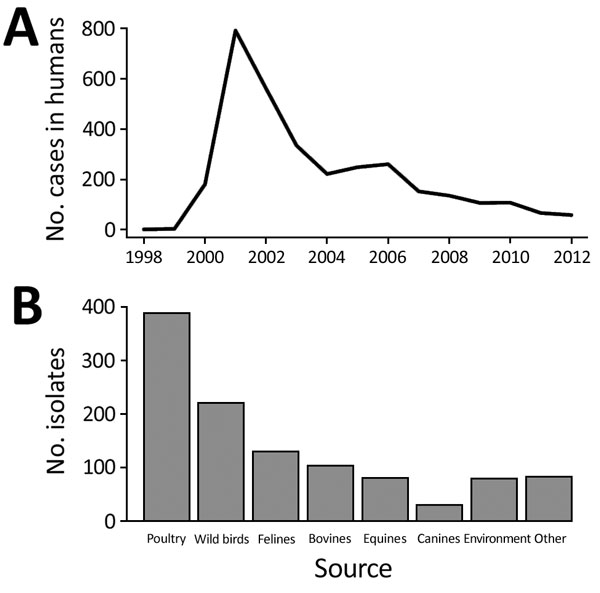
Figure 1
References
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al.; International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–9. DOIPubMedGoogle Scholar
- Institute of Environmental Science and Research Ltd. (ESR). Notifiable diseases. New Zealand public health observatory. 2014 [cited 2016 Feb 2]. http://www.nzpho.org.nz/NotifiableDisease.aspx
- King N, Lake R, Campbell D. Source attribution of nontyphoid salmonellosis in new zealand using outbreak surveillance data. J Food Prot. 2011;74:438–45. DOIPubMedGoogle Scholar
- Eswarappa SM, Janice J, Nagarajan AG, Balasundaram SV, Karnam G, Dixit NM, et al. Differentially evolved genes of Salmonella pathogenicity islands: insights into the mechanism of host specificity in Salmonella. PLoS One. 2008;3:e3829. DOIPubMedGoogle Scholar
- Lal A, Baker MG, French NP, Dufour M, Hales S. The epidemiology of human salmonellosis in New Zealand, 1997-2008. Epidemiol Infect. 2012;140:1685–94. DOIPubMedGoogle Scholar
- Lal A, Ikeda T, French N, Baker MG, Hales S. Climate variability, weather and enteric disease incidence in New Zealand: time series analysis. PLoS One. 2013;8:e83484. DOIPubMedGoogle Scholar
- Alley MR, Connolly JH, Fenwick SG, Mackereth GF, Leyland MJ, Rogers LE, et al. An epidemic of salmonellosis caused by Salmonella Typhimurium DT160 in wild birds and humans in New Zealand. N Z Vet J. 2002;50:170–6. DOIPubMedGoogle Scholar
- Institute of Environmental Science and Research Ltd (ESR). ESR LabLink. Quarterly surveillance summaries for New Zealand, March 2000–March 2003 [cited 2016 Nov 25]. https://surv.esr.cri.nz/PDF_surveillance/Lablink/
- Institute of Environmental Science and Research Ltd (ESR). Public Health Surveillance; Information for New Zealand public health action. 2003–2012 human Salmonella isolates [cited 2016 Nov 25]. https://surv.esr.cri.nz/enteric_reference/human_salmonella.php
- Institute of Environmental Science and Research Ltd (ESR). Public Health Surveillance; Information for New Zealand public health action. Non-human Salmonella isolates, 2003–2012 [cited 2016 Nov 25]. https://surv.esr.cri.nz/enteric_reference/nonhuman_salmonella.php
- Penfold JB, Amery HC, Peet PJ. Gastroenteritis associated with wild birds in a hospital kitchen. Br Med J. 1979;2:802.PubMedGoogle Scholar
- Tizard IR, Fish NA, Harmeson J. Free flying sparrows as carriers of salmonellosis. Can Vet J. 1979;20:143–4.PubMedGoogle Scholar
- Lawson B, Howard T, Kirkwood JK, Macgregor SK, Perkins M, Robinson RA, et al. Epidemiology of salmonellosis in garden birds in England and Wales, 1993 to 2003. EcoHealth. 2010;7:294–306. DOIPubMedGoogle Scholar
- Piccirillo A, Mazzariol S, Caliari D, Menandro ML. Salmonella Typhimurium phage type DT160 infection in two Moluccan cockatoos (Cacatua moluccensis): clinical presentation and pathology. Avian Dis. 2010;54:131–5. DOIPubMedGoogle Scholar
- Grillo T, Post L. Salmonella Typhimurium DT160 outbreak in Tasmania. Animal Health Surveillance Quarterly Reports. 2010;14:8–8. http://www.sciquest.org.nz/node/72986
- QIAGEN. QIAamp DNA mini and blood mini handbook. Third edition. 2012 [cited 2015 Feb 12]. https://moodle.ufsc.br/pluginfile.php/1379318/mod_resource/content/0/QIAamp_DNA_Mini_Blood.pdf
- Aronesty E. Comparison of sequencing utility programs. Open Bioinform J. 2013;7:1–8. DOIGoogle Scholar
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. DOIPubMedGoogle Scholar
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5. DOIPubMedGoogle Scholar
- Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–8. DOIPubMedGoogle Scholar
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. DOIPubMedGoogle Scholar
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. DOIPubMedGoogle Scholar
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 [cited 2017 Feb 2]. https://arxiv.org/abs/1207.3907
- Petrovska L, Mather AE, AbuOun M, Branchu P, Harris SR, Connor T, et al. Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005–2010. Emerg Infect Dis. 2016;22:617–24. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- He Z, Zhang H, Gao S, Lercher MJ, Chen WH, Hu S. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44(W1):W236-41. DOIPubMedGoogle Scholar
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–5. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. DOIPubMedGoogle Scholar
- Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol. 2008;25:1459–71. DOIPubMedGoogle Scholar
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. DOIPubMedGoogle Scholar
- Rambaut A, Suchard MA, Xie D, Drummond AJ. Beast. Tracer 1.6 2014 [cited 2016 Sep 27]. http://beast.bio.ed.ac.uk/Tracer
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5:e1000520. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. DOIPubMedGoogle Scholar
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–6. DOIPubMedGoogle Scholar
- Clark KR, Gorle RN. PRIMER v6: user manual/tutorial. Plymouth (UK): PRIMER-E; 2006. p. 296.
- Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62:245–53. DOIPubMedGoogle Scholar
- Popoff MY, Le Minor LE. Genus XXXIII. Salmonella. In: Brenner DJ, Staley JT, editors. Bergey’s manual of systematic bacteriology. New York: Springer; 2005. p. 764–99.
- Thornley CN, Simmons GC, Callaghan ML, Nicol CM, Baker MG, Gilmore KS, et al. First incursion of Salmonella enterica serotype typhimurium DT160 into New Zealand. Emerg Infect Dis. 2003;9:493–5. DOIPubMedGoogle Scholar
- Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, Harris SR, et al. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science. 2013;341:1514–7. DOIPubMedGoogle Scholar
- Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–21. DOIPubMedGoogle Scholar
- Gieraltowski L, Julian E, Pringle J, Macdonald K, Quilliam D, Marsden-Haug N, et al. Nationwide outbreak of Salmonella Montevideo infections associated with contaminated imported black and red pepper: warehouse membership cards provide critical clues to identify the source. Epidemiol Infect. 2013;141:1244–52. DOIPubMedGoogle Scholar
- Byrne L, Fisher I, Peters T, Mather A, Thomson N, Rosner B, et al.; International Outbreak Control Team. A multi-country outbreak of Salmonella Newport gastroenteritis in Europe associated with watermelon from Brazil, confirmed by whole genome sequencing: October 2011 to January 2012. Euro Surveill. 2014;19:6–13. DOIPubMedGoogle Scholar
- Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, Tavazoie S. Bacterial adaptation through loss of function. PLoS Genet. 2013;9:e1003617. DOIPubMedGoogle Scholar
- Koskiniemi S, Sun S, Berg OG, Andersson DI. Selection-driven gene loss in bacteria. PLoS Genet. 2012;8:e1002787. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.
Page created: May 16, 2017
Page updated: May 16, 2017
Page reviewed: May 16, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.