Volume 24, Number 11—November 2018
Dispatch
Emergence of Neisseria meningitidis Serogroup W, Central African Republic, 2015–2016
Abstract
We analyzed data from the 2015 and 2016 meningitis epidemic seasons in Central African Republic as part of the national disease surveillance. Of 80 tested specimens, 66 belonged to meningococcal serogroup W. Further analysis found that 97.7% of 44 isolates belonged to the hyperinvasive clonal complex sequence type 11.
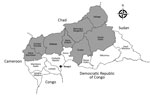
Central African Republic (CAR) is localized in the meningitis belt that spans from Senegal to Ethiopia in sub-Saharan Africa. The country experiences meningitis epidemics every year. Eight health districts (Nana Mambéré, Ouham Pendé, Ouham, Nana Gribizi, Bamingui Bangoran, Vakaga, Haute-Kotto, and Ouaka) are at particularly high risk of meningitis outbreaks during the dry season, November–March (Figure 1). During this period, epidemiologic surveillance is reinforced by the Health Ministry with the help of the World Health Organization (WHO), Institut Pasteur de Bangui (IPB; Bangui, CAR), and nongovernmental organizations, depending on the geographic zone.
The gram-negative bacterium Neisseria meningitidis remains the leading cause of meningitis epidemics in Africa. Because of political troubles in the past years and missing reliable information available from the provinces, few epidemiologic and biologic data are available in CAR. Two previous reports, published by IPB in 2006 and 2008, had focused only on cases in Bangui (1,2). Data showed that, among proven bacterial disease, Streptococcus pneumoniae was the most common organism responsible for meningitis, resulting in a 47% death rate (29/62 cases). N. meningitidis, especially serogroup A (NmA), was also responsible for menigitis in Bangui to a lesser extent (1,2).
Historically, NmA of sequence types (STs) 5 and 7 was predominant in sub–Saharan Africa; serogroups W (ST11), C, and X (ST181) caused smaller numbers of cases. After introduction of massive vaccination campaigns, serogroup A incidence has massively decreased, and meningococcal strains causing disease shifted toward non-A serogroups (3). In particular, after the 1996–1997 epidemic in Africa that killed nearly 250,000 persons, the introduction of the MenAfriVac vaccine in 2010 contributed to dramatically reduced NmA cases in 26 countries of the meningitis belt by targeting roughly 65 million persons 1–29 years of age (WHO, http://www.who.int/immunization/newsroom/menafrivac_20121114/en/). Consequently, serogroup replacement has been noted. For example, in 2015, an epidemic with a novel strain of serogroup C was recorded in Niger and Nigeria for the first time since 1975 (WHO, http://www.who.int/wer/2015/wer9047.pdf?ua=1).
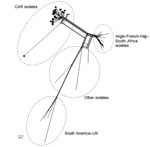
Hyperinvasive N. meningitidis serogroup W (NmW) cases belonging to the clonal complex ST11 (NmW/cc11) have been reported worldwide since the Hajj 2000–linked outbreak (3–6). Moreover, NmW/cc11 cases have been reported in the African meningitis belt at least since the late 1990s (7). Epidemics caused by these isolates were reported in 2001 and declined thereafter, but NmW/cc11 seems to have reemerged since 2010 (8). The genetic relationships between these NmW/cc11 isolates have been recently resolved using whole-genome sequencing, and 2 major NmW/cc11 lineages were described—the Anglo-French Hajj and South American/UK strains—in addition to several local NmW/cc11 isolates (5). We aimed to explore the genetic relationships of the 2015 and 2016 isolates from CAR with this reported population structure of NmW/cc11.
This study was a collaborative work led by IPB. In 2015 and 2016, CAR experienced annual national meningitis epidemics. All cases confirmed positive for meningococcal meningitis in IPB were from 7 districts in Ouham Prefecture: Batangafo, Besse, Lady, Kambakota, Ouogo, Bongonon, and Kabo.
Each suspected patient meeting the recommended WHO case definition for meningitis (sudden onset of fever >38.5°C rectal or >38.0°C axillary and 1 of the following signs: neck stiffness, altered consciousness, or other meningeal sign) underwent a lumbar puncture and received antimicrobial drug therapy. In some cases, when available, a latex agglutination test (Pastorex; Bio-Rad, Hercules, CA, USA) was done on cerebrospinal fluid (CSF) samples before an aliquot and a clinical case report form for each patient were transported to IPB. We analyzed each sample according to procedures routinely used in IPB for diagnosis and monitoring of meningitis, according to WHO methodology (http://www.who.int/csr/resources/publications/meningitis/WHO_CDS_CSR_EDC_99_7_EN/en/). In brief, laboratory methodology used for NmW identification included Gram stain procedure, primary culture of CSF or transisolate inoculated medium onto blood agar plate and chocolate agar, and PCR testing. If CSF was visibly cloudy when received, we also performed latex agglutination (Pastorex) in the laboratory. NmW was considered as the causative agent when identified by culture, PCR, or both.
We performed molecular identification and genogroup determination using multiplex PCR (9,10) and multilocus sequence typing (MLST) using the methodology of the PubMLST Neisseria database (http://pubmlst.org/neisseria) on CSF samples (n = 44) using a single round of PCR. We subjected a proportion of cultured isolates (n = 28) to whole-genome sequencing using Illumina NextSeq 500 (Illumina, San Diego, CA, USA) and assembled them as described previously (11). We compared genomes using the PubMLST Genome Comparator tool on the basis of core genome (12). We visualized phylogenetic trees using SplitsTree4 version 4.13.1 (http://www.splitstree.org).
In 2015 and 2016, a total of 276 suspected cases of meningitis were reported in CAR (Table). Among these 276 patients, 13 died in 2015 and 12 in 2016. Of the 80 CSF samples received at IPB for biologic investigations, 52 bacteriologic cultures and 66 PCR tests were positive for NmW. All samples that were positive in culture were also positive in PCR. All 66 positive PCR tests identified serogroup W, confirming the presence of such epidemic strains in central Africa. We also found 5 PCR tests positive for Streptococcus pneumoniae, as well as 9 negative results for both culture and PCR assays on the CSF samples. The most common signs and symptoms among the confirmed NmW case-patients were fever (97%), weakness (85%), and neck stiffness (78%).
From the 66 confirmed NmW cases, 44 CSF samples were sent to the WHO collaborating reference center for meningitis located in Institut Pasteur, Paris, for diagnostic confirmation and genetic analyses. Additionally, 28 isolates underwent genome sequence analysis for evaluating the genetic relationships of NmW strains identified in CAR to others during epidemics elsewhere. Phylogeny results clearly indicated that all the CAR isolates that underwent next-generation sequencing (4 in 2015 and 24 in 2016) were genetically linked. These isolates grouped together in a genetic cluster separated from other NmW/cc11 strains previously described, including the Anglo-French Hajj branch and the South American/UK branch, and from other local strains found in Africa (Figure 2).
Compared with data obtained in Bangui in the past years, which highlighted NmA as the main etiology of meningitis cases, our data shows that the strains isolated during epidemic seasons in the north of CAR in 2015 and 2016 were all NmW. In addition, most of the characterized cases (43/44) belonged to the ST11 complex and had the antigenic formula NmW:P1.5,2:F1–1:cc11. The other case isolated in 2016 (1/44) belonged to the ST175 complex, a nonhyperinvasive clonal complex that should promote immune exploration, as such isolates have been reported more frequently among patients with complement deficiencies (13).
The number of meningitis cases reported during the 2015 and 2016 epidemic seasons was the most documented in CAR since 2008. We described the presence of a specific endemic NmW strain that appears to be circulating in sub–Saharan Africa, including in CAR. Foci correspond to endemic local strains, which confirms the hypothesis of the multifocal emergence of specific NmW/cc11 strains (5,14,15).
Although CAR experiences epidemics of meningitis each year, the weaknesses of the national health system contribute to a delay in management of suspected cases. The emergence or reemergence of epidemic NmW strains in CAR requires persistent public health surveillance and increased capacity of epidemic detection and prevention, as well as revised vaccination policy.
Dr. Frank is a senior medical microbiologist in the Laboratory for Biomedical Analysis of the Institut Pasteur, Bangui, Central African Republic. His primary research interests include surveillance of bacterial diseases such as meningitis in Africa, as well as antimicrobial drug resistance.
Acknowledgments
We thank the health ministry and local health authorities of Central African Republic. We gratefully acknowledge all the health district staff involved in management of meningitis epidemics, as well as Doctors without Borders from Spain, for constant collaboration and procurement of samples during outbreaks in Central African Republic. We also thank Alexandre Manirakiza and the Epidemiology Department of IPB for having created and customized the map in Figure 1.
This work was supported by the French Ministry of Foreign Affairs and Institut Pasteur, Bangui, Central African Republic, and Paris, France.
References
- Békondi C, Bernede C, Passone N, Minssart P, Kamalo C, Mbolidi D, et al. Primary and opportunistic pathogens associated with meningitis in adults in Bangui, Central African Republic, in relation to human immunodeficiency virus serostatus. Int J Infect Dis. 2006;10:387–95. DOIPubMedGoogle Scholar
- Bercion R, Bobossi-Serengbe G, Gody JC, Beyam EN, Manirakiza A, Le Faou A. Acute bacterial meningitis at the ‘Complexe Pédiatrique’ of Bangui, Central African Republic. J Trop Pediatr. 2008;54:125–8. DOIPubMedGoogle Scholar
- Mustapha MM, Marsh JW, Harrison LH. Global epidemiology of capsular group W meningococcal disease (1970-2015): Multifocal emergence and persistence of hypervirulent sequence type (ST)-11 clonal complex. Vaccine. 2016;34:1515–23. DOIPubMedGoogle Scholar
- Ladhani SN, Beebeejaun K, Lucidarme J, Campbell H, Gray S, Kaczmarski E, et al. Increase in endemic Neisseria meningitidis capsular group W sequence type 11 complex associated with severe invasive disease in England and Wales. Clin Infect Dis. 2015;60:578–85. DOIPubMedGoogle Scholar
- Lucidarme J, Hill DM, Bratcher HB, Gray SJ, du Plessis M, Tsang RS, et al. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect. 2015;71:544–52. DOIPubMedGoogle Scholar
- Zhou H, Liu W, Xu L, Deng L, Deng Q, Zhuo J, et al. Spread of Neisseria meningitidis serogroup W clone, China. Emerg Infect Dis. 2013;19:1496–9. DOIPubMedGoogle Scholar
- Kwara A, Adegbola RA, Corrah PT, Weber M, Achtman M, Morelli G, et al. Meningitis caused by a serogroup W135 clone of the ET-37 complex of Neisseria meningitidis in West Africa. Trop Med Int Health. 1998;3:742–6. DOIPubMedGoogle Scholar
- Collard JM, Maman Z, Yacouba H, Djibo S, Nicolas P, Jusot JF, et al. Increase in Neisseria meningitidis serogroup W135, Niger, 2010. Emerg Infect Dis. 2010;16:1496–8. DOIPubMedGoogle Scholar
- Fraisier C, Stor R, Tenebray B, Sanson Y, Nicolas P. Use of a new single multiplex PCR-based assay for direct simultaneous characterization of six Neisseria meningitidis serogroups. J Clin Microbiol. 2009;47:2662–6. DOIPubMedGoogle Scholar
- Bronska E, Kalmusova J, Dzupova O, Maresova V, Kriz P, Benes J. Dynamics of PCR-based diagnosis in patients with invasive meningococcal disease. Clin Microbiol Infect. 2006;12:137–41. DOIPubMedGoogle Scholar
- Taha MK, Claus H, Lappann M, Veyrier FJ, Otto A, Becher D, et al. Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLoS One. 2016;11:e0154047. DOIPubMedGoogle Scholar
- Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. DOIPubMedGoogle Scholar
- Rosain J, Hong E, Fieschi C, Martins PV, El Sissy C, Deghmane AE, et al. Strains responsible for invasive meningococcal disease in patients with terminal complement pathway deficiencies. J Infect Dis. 2017;215:1331–8. DOIPubMedGoogle Scholar
- Hossain MJ, Roca A, Mackenzie GA, Jasseh M, Hossain MI, Muhammad S, et al. Serogroup W135 meningococcal disease, The Gambia, 2012. Emerg Infect Dis. 2013;19:1507–10. DOIPubMedGoogle Scholar
- Cibrelus L, Medah I, Koussoubé D, Yélbeogo D, Fernandez K, Lingani C, et al. Serogroup W meningitis outbreak at the subdistrict level, Burkina Faso, 2012. Emerg Infect Dis. 2015;21:2063–6. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: September 21, 2018
1These authors contributed equally to this article.
Table of Contents – Volume 24, Number 11—November 2018
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Pierre-Alain Rubbo, Institut Pasteur de Bangui, BP 923, Avenue de l’Indépendance, Bangui, Central African Republic
Top