Volume 25, Number 12—December 2019
Dispatch
Distantly Related Rotaviruses in Common Shrews, Germany, 2004–2014
Figure 2
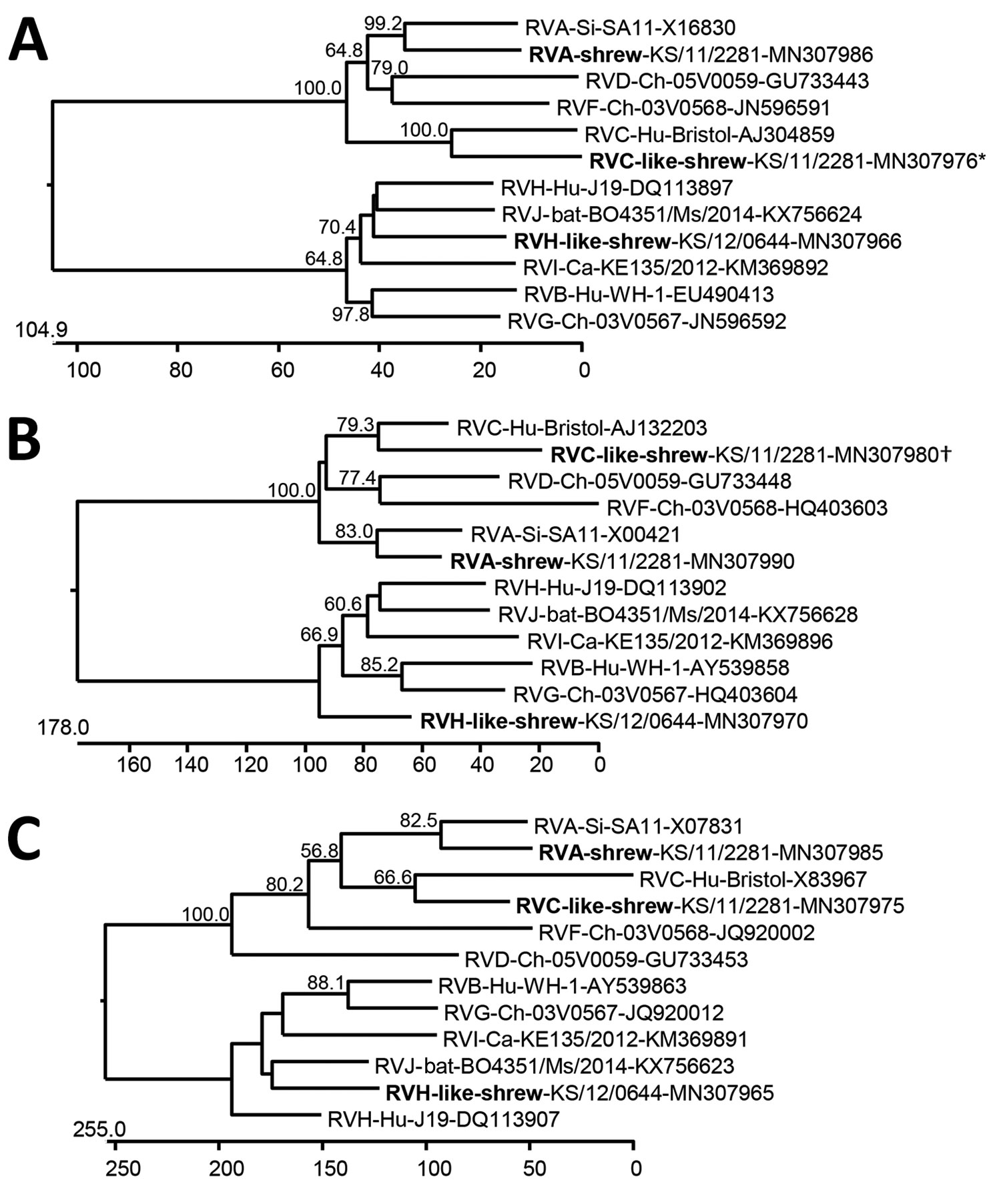
Figure 2. Phylogenetic relationship of shrew rotaviruses (bold), Germany, 2011–2012, with RVA–RVJ determined by using the deduced amino acid sequences of virus protein 1 (A), virus protein 6 (B), and nonstructural protein 5 (C). Trees were constructed by using a neighbor-joining method implemented in the MegAlign module of DNASTAR (https://www.dnastar.com) and a bootstrap analysis with 1,000 trials and 111 random seeds. Bootstrap values of >50% are shown. The rotavirus species, host, strain or sample designation, and GenBank accession number are indicated at each branch. Scale bars indicate amino acid substitutions per 100 residues. *Sequence incomplete at N terminus (≈70 aa residues missing) and C terminus (≈10 aa residues missing). †Sequence incomplete at N terminus (≈40 aa residues missing). Ca, canine; Ch, chicken; Hu, human; RVA, rotavirus A; RVB, rotavirus B; RVC, rotavirus C; RVD, rotavirus D; RVF, rotavirus F; RVG, rotavirus G; RVH, rotavirus H; RVI, rotavirus I; RVJ, rotavirus J; Si, simian.