Volume 25, Number 3—March 2019
Research Letter
Exportation of MDR TB to Europe from Setting with Actively Transmitted Persistent Strains in Peru
Abstract
We performed a cross-border molecular epidemiology analysis of multidrug-resistant tuberculosis in Peru, Spain, and Italy. This analysis revealed frequent transmission in Peru and exportation of a strain that recreated similar levels of transmission in Europe during 2007–2017. Transnational efforts are needed to control transmission of multidrug-resistant tuberculosis globally.
International migratory movements have created a need for cross-border surveillance of tuberculosis (TB). Monitoring the transmission of multidrug-resistant (MDR) Mycobacterium tuberculosis strains deserves further analysis (1). Through migration, MDR strains can become more widely dispersed; they can be exported from the 30 countries with 89.7% of the incident MDR cases (2) to lower prevalence settings.
We performed a transnational molecular epidemiology analysis of MDR TB cases covering a setting with one of the highest resistance rates in Latin America (Lima, Peru) (2) and 2 settings in Europe hosting immigrants from Peru (Florence, Italy; and Madrid, Spain) to identify incidents of cross-border transmission. We selected 60 consecutive MDR TB cases (20% of the total MDR cases in Lima) diagnosed during 2014–2015 in one of the poorest districts of Lima (San Juan de Lurigancho), which has the highest incidence of TB (193 cases/100,000 population) in Peru (3). MIRU-VNTR (mycobacterial interspersed repetitive unit–variable-number tandem-repeat) analysis (Appendix 1) suggested a high percentage of recent transmission that included 36 (60%) of 60 isolates in 9 clusters (Appendix 2). A comparison of these isolates with 228 genotyped isolates from the same district 4 years earlier (3) revealed that 6 of the 9 strains actively transmitted during 2014–2015 were present in 2011 (Appendix 1 Table 1).
We then investigated whether some of these persistent MDR TB strains actively transmitted in Lima could have been exported to Europe. We used a dataset of 87 MIRU-VNTR genotypes of isolates in Florence obtained from TB cases in Peru during 2001–2010 (4) and >300 MDR genotyped isolates obtained nationwide from Italy (5). We found that 1 genotype matched between the Lima and Italy MDR datasets; this genotype corresponded to a strain (C8-LPMDR) that infected 11 persons in Florence and 2 in Milan during 2007–2017 (Appendix 1 Table 2). MDR TB strains from Lima were also found in Spain during 2003–2009. Three MDR isolates, matching 3 of the 9 MDR TB strains from Lima, were found in migrants from Peru residing in Madrid (Appendix 1 Table 1). One of these isolates corresponded to the active MDR strain circulating in Italy (C8-LPMDR).
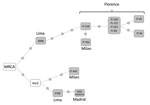
We performed whole-genome sequencing (6) with 12 of the 17 isolates of the cross-border MDR TB cluster C8-LPMDR (7 from Florence, 2 from Milan, 2 from Lima, and 1 from Madrid). In a median-joining network analysis, these isolates were distributed along 2 branches (Figure). One branch included all the isolates from Florence. Although we lacked precise data from contact tracing to verify details regarding transmission in Florence, we were able to determine that all the Peru migrants involved came from Lima. In Florence, there is a large community of persons from Peru, which offers opportunities for interacting, such as shared residence and social gatherings. The few differences (0–2 single-nucleotide polymorphisms [SNPs]) found among these isolates strongly suggests these isolates were recently transmitted in Florence. An isolate from Lima (6068) was only 3 SNPs different from a Florence isolate, demonstrating a close genetic relationship between the Florence and Lima isolates. This close relationship also suggests that the starting point of this branch was an exportation event of an isolate from Lima. The second branch in the network includes 2 isolates identified in Europe (1 Madrid [city of origin unknown, data not available] and 1 Milan [origin Lima]) and 1 isolate identified in Lima. Because the most recent common ancestor is positioned between the 2 branches and the 2 isolates from Lima are in different branches, these branches probably represent 2 independent exportations of 2 variants of a strain prevalent in Lima that diversified after a prolonged period.
These data reveal that high-risk strains are being exported from Lima to 2 countries of Europe (Italy and Spain). Not only were these strains exported from Lima, but 1 strain caused a prolonged and ongoing transmission event in Italy. The transmission of this strain has caused at least 3 cases in Lima, 11 in Florence, 2 in Milan, and 1 in Madrid.
In another report, the international distribution of an MDR TB strain that caused 10 cases across 3 countries of Europe (Romania, Austria, and Germany) was investigated (7). The exportation event discussed in our report is geographically wider (intercontinental, from South America to Europe), involved more cases (17 total, with a transmission cluster of 12 cases in Italy), and occurred over a more extended period (secondary cases spanned 11 years).
Only integrative transnational efforts can provide a clearer picture of transmission of MDR TB, which has become more complex because of international migration. In this cooperative analysis involving Peru, Italy, and Spain, we detected a serious problem of active MDR TB transmission in Lima. This situation led to a pool of persistent strains that were responsible for similar transmission events after exportation to Europe via migration.
Mr. Acosta is an investigator at the Instituto de Investigación Sanitaria Gregorio Marañón and the Servicio de Microbiología of the Hospital General Universitario Gregorio Marañón in Madrid, Spain. His research interests include TB transmission and developing simplified tools to optimize tracking of transmission.
Acknowledgments
We thank Thomas O’Boyle for proofreading the manuscript.
This project was funded by ERANet-LAC (grant nos. ELAC2015/T08-0664, E035-ERAnet-LAC/J110-2016/FONDECYT, PER-2012-ELAC2015/T08-0664) and the Instituto de Salud Carlos III (grant nos. AC16/00057, FIS15/01554, 13/01207, 16/01449). L.P.-L. holds a Miguel Servet grant (no. CP15/00075). F.A. holds a grant from the Institute for the Formation and Use of Human Resources, National Secretariat of Science and Technology of Panama (no. 270-2016-293).
References
- Zignol M, Dean AS, Falzon D, van Gemert W, Wright A, van Deun A, et al. Twenty years of global surveillance of antituberculosis-drug resistance. N Engl J Med. 2016;375:1081–9. DOIPubMedGoogle Scholar
- World Health Organization. Global tuberculosis report 2017. 2017 [cited 2018 Sep 28]. http://apps.who.int/medicinedocs/ documents/s23360en/s23360en.pdf
- Barletta F, Otero L, de Jong BC, Iwamoto T, Arikawa K, Van der Stuyft P, et al. Predominant Mycobacterium tuberculosis families and high rates of recent transmission among new cases are not associated with primary multidrug resistance in Lima, Peru. J Clin Microbiol. 2015;53:1854–63. DOIPubMedGoogle Scholar
- Zammarchi L, Tortoli E, Borroni E, Bartalesi F, Strohmeyer M, Baretti S, et al.; COHEMI project study group. High prevalence of clustered tuberculosis cases in peruvian migrants in florence, Italy. Infect Dis Rep. 2014;6:5646. DOIPubMedGoogle Scholar
- European Centre for Disease Prevention and Control. Molecular typing for surveillance of multidrug-resistant tuberculosis in the EU/EEA. 2017 Mar 6 [cited 2018 Sep 28]. https://ecdc.europa.eu/en/publications-data/molecular-typing-surveillance-multidrug-resistant-tuberculosis-eueea
- Herranz M, Pole I, Ozere I, Chiner-Oms Á, Martínez-Lirola M, Pérez-García F, et al. Mycobacterium tuberculosis acquires limited genetic diversity in prolonged infections, reactivations and transmissions involving multiple hosts. Front Microbiol. 2018;8:2661. DOIPubMedGoogle Scholar
- Fiebig L, Kohl TA, Popovici O, Mühlenfeld M, Indra A, Homorodean D, et al. A joint cross-border investigation of a cluster of multidrug-resistant tuberculosis in Austria, Romania and Germany in 2014 using classic, genotyping and whole genome sequencing methods: lessons learnt. Euro Surveill. 2017;22:30439. DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: January 31, 2019
Table of Contents – Volume 25, Number 3—March 2019
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Darío García de Viedma, Hospital Gregorio Marañón, Servicio de Microbiologia, C, Dr Esquerdo 46, Madrid 28007, Spain
Top