Volume 26, Number 6—June 2020
Research
Antimicrobial Resistance in Salmonella enterica Serovar Paratyphi B Variant Java in Poultry from Europe and Latin America
Figure 3
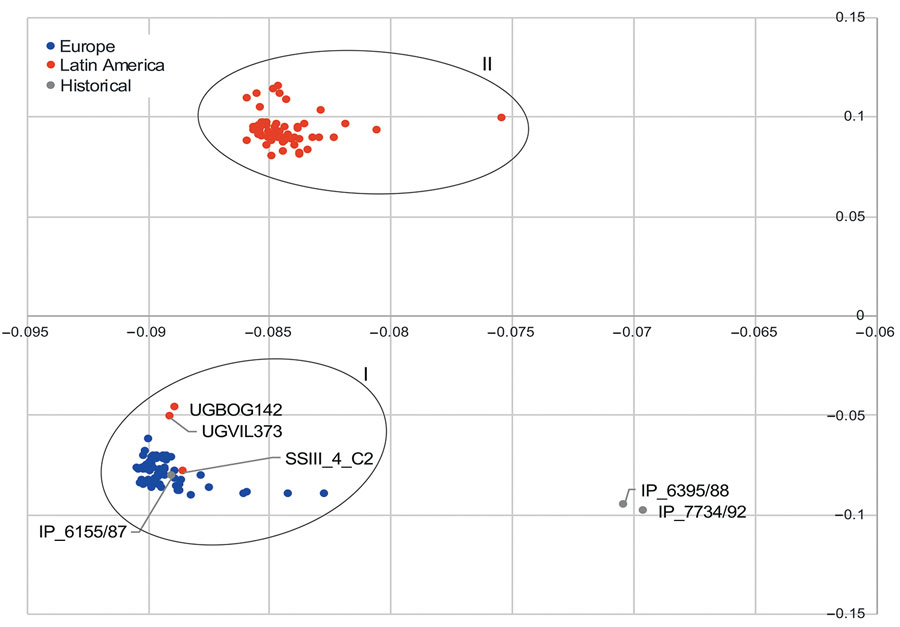
Figure 3. Principal component analysis plot comparing accessory (noncore) genome of chromosome contigs of strains of Salmonella enterica serovar Paratyphi B variant Java sequence type 28. Oval rings indicate clusters I and II. Cluster I grouped together historical strain IP_6155/87 with all strains from Europe and some from Latin American. Cluster II grouped Latin America strains only. Cluster II was associated with a prophage sequence highly similar to the Salmonella phage SEN34 (National Center for Biotechnology Information reference sequence no. NC_028699.1). The prophage sequence was absent in strains from cluster I.
1Current affiliate: Quadram Institute Bioscience, Norwich, UK.
2These authors contributed equally to this article.
Page created: May 18, 2020
Page updated: May 18, 2020
Page reviewed: May 18, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.