Volume 28, Number 1—January 2022
Research
High-Level Quinolone-Resistant Haemophilus haemolyticus in Pediatric Patient with No History of Quinolone Exposure
Abstract
The prevalence of antimicrobial resistance among Haemophilus spp. is a critical concern, but high-level quinolone-resistant strains had not been isolated from children. We isolated high-level quinolone-resistant H. haemolyticus from the suction sputum of a 9-year-old patient. The patient had received home medical care with mechanical ventilation for 2 years and had not been exposed to any quinolones for >3 years. The H. haemolyticus strain we isolated, 2019-19, shared biochemical features with H. influenzae. However, whole-genome analysis found this strain was closer to H. haemolyticus. Phylogenetic and mass spectrometry analyses indicated that strain 2019-19 was in the same cluster as H. haemolyticus. Comparison of quinolone resistance–determining regions showed strain 2019-19 possessed various amino acid substitutions, including those associated with quinolone resistance. This report highlights the existence of high-level quinolone-resistant Haemophilus species that have been isolated from both adults and children.
The genus Haemophilus includes 9 bacterial species that cause infections only among humans: H. influenzae, H. aegyptius, H. haemolyticus, H. parainfluenzae, H. parahaemolyticus, H. paraphrohaemolyticus, H. pittmaniae, H. sputorum, and H. ducreyi (1). Among them, H. influenzae is a notable causative pathogen of respiratory infections and otitis media in children (2–4). H. haemolyticus, which is genetically similar to H. influenzae and coexists with H. influenzae in the upper respiratory tract (1,5), is considered a commensal bacterium, and its pathogenicity has not been widely examined. However, some previous studies have reported that H. haemolyticus can be misidentified as H. influenzae in the clinical setting (6–8).
β-lactams and quinolones are commonly used to treat infections caused by both H. influenzae and H. haemolyticus. Recently, an increase in H. influenzae strains with reduced susceptibility to quinolones has been reported (9–11). Moreover, high-level resistant strains (MIC for levofloxacin ≥8 μg/mL) of H. influenzae have also emerged (12–15) but have been isolated only from adult case-patients (11,12). Recent nationwide surveillance in Japan indicated that quinolone-resistant H. influenzae had not been isolated among pediatric patients (16). Although low-susceptibility strains of H. haemolyticus have emerged, a high-level resistance strain had not been isolated from a pediatric patient (17). We isolated the H. haemolyticus strain 2019-19, which showed high-level resistance to quinolones, from a pediatric patient in an acute care hospital in Tokyo, Japan, and analyzed the features of the strain and case background of the patient.
Patient Characteristics
A 9-year-old girl with severe motor and intellectual disabilities, hypothyroidism, and chronic respiratory disease was hospitalized for hypoxic ischemic encephalopathy in the Tokyo University of Hachioji Medical Centre (Tokyo, Japan). The patient had been under mechanical ventilation related to tracheostomy since she was 7 years of age and had not been administered any quinolones for >3 years before hospitalization. Because the patient had a fever with increased sputum production after 3 days of hospitalization, we obtained a suction sputum culture and administered ampicillin/sulbactam intravenously to her for 1 week. The patient was discharged because her fever resolved after 5 days of hospitalization.
Bacterial Isolation and Culture Conditions
We isolated H. haemolyticus (strain identification 2019-19) from the suction sputum and identified it as quinolone-resistant H. influenzae by routine laboratory testing using a MicroScan WalkAway system (Siemens, https://www.siemens.com). Because quinolone-resistant H. influenzae had never been isolated from a pediatric patient, we performed a detailed susceptibility test for 2019-19 by the broth microdilution method. For controls in the biochemical test, we used H. influenzae GTC 14202T (DSM 4690T) and H. haemolyticus GTC 15009T (NCTC 10659T) type strains purchased from Gifu University (https://www.gifu-u.ac.jp). In addition, we used H. influenzae ATCC 49247 and Rd as quality control strains for antimicrobial susceptibility testing. We cultured the isolates overnight on chocolate agar at 37°C in a 5% CO2 atmosphere and stored them in 10% skim milk at −80°C until use. This study was approved by the research ethics committees at the Tokyo University of Pharmacy and Life Sciences (case no. 16-12).
Antimicrobial Susceptibility Test
We measured MICs by broth microdilution method as described by the Clinical and Laboratory Standards Institute (18). As tested agents, we used ampicillin, amoxicillin, clavulanic acid, cefotaxime, meropenem, clarithromycin, azithromycin, levofloxacin, tosufloxacin, and moxifloxacin. In addition, we used PAβN (Phe-Arg β-naphthylamide dihydro-chloride; Sigma-Aldrich; https://www.sigmaaldrich.com) and reserpine (Sigma-Aldrich) as efflux pump inhibitors. We set antimicrobial-susceptible breakpoints according to Clinical and Laboratory Standard Institute criteria (18).
Genomic Analysis
We extracted genomic DNA using a Wizard Genomic DNA purification kit (Promega, https://www.promega.com) and sequenced it using GridION (Oxford Nanopore Technologies, https://nanoporetech.com) and DNB Seq-G400 (MGI Tech, https://en.mgi-tech.com) according to manufacturer instructions. We assembled the sequenced data with Unicycler version 0.4.7 (https://github.com/rrwick/Unicycler) with default parameters, assessed the quality of the genome using CheckM, version 1.0.12 (https://github.com/Ecogenomics/CheckM), and annotated the assembled genome sequence using DDBJ’s DFAST Fast Annotation and Submission Tool; https://dfast.nig.ac.jp). The obtained and annotated sequence data were registered in the DDBJ database under DDBJ/EMBL/GenBank accession number AP024093.
Because the genome sequence of the type strain was not available, we used H. haemolyticus NCTC 10839, along with H. influenzae ATCC 33391T, to compare the entire genomes with strain 2019-19, using Easyfig version 2.2.2 (19). We calculated the average nucleotide identity (ANIb) algorithm using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with JSpeciesWS (20) and digital DNA-DNA hybridization (dDDH) using the type strain genome server (21). In addition, we estimated the presence of CRISPR sequences and cas genes in the genome using the CRISPRfinder program (22).
Phylogenetic Analysis of Typical Genes
We illustrated a phylogenetic dendrogram using typical genes (16S rDNA sequence, adk, pgi, recA, infB, gyrA, gyrB, parC, parE, and ftsI) with Clustal Omega alignment and the neighbor-joining method of Jukes-Cantor using Geneious Prime 2019 (Biomatters, https://www.geneious.com). We selected the 16S rDNA sequence, adk, pgi, recA, and infB because they were used in a previous classification study (1,23). In addition, we used gyrA, gyrB, parC, parE, and ftsI as antimicrobial-targeting genes. We used nucleotide sequences of Escherichia coli ATCC 11775T as an outgroup.
Biochemical Test
We used an API NH kit (bioMérieux, https://www.biomerieux.com) to assay biochemical characteristics, prepare bacterial cultures, and interpret the results according to manufacturer protocols. We evaluated use of the V factor, X factor, and several nutrients using Haemophilus ID Quad with growth factors agar (BD Biosciences, https://www.bdbiosciences.com). We cultured the agar plates overnight at various temperatures (4°C, 16°C, 25°C, 37°C, and 42°C).
Mass Spectrometry
We prepared samples by ethanol/formic acid extraction. One loop of bacteria was suspended in 300 μL distilled water and 900 μL of ethanol was added into the suspension. After centrifuging and discarding the supernatant, we mixed 20 μL each of 70% formic acid and acetonitrile. Next, we applied 1 μL of supernatant on the target plate and mixed it with 1 μL HCCA matrix (Bruker, https://www.bruker.com). We obtained the spectrum using a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS Bruker autoflex maX and analyzed the phylogenetic tree using MALDI Biotyper Compass Explorer version 4.1.60 (Bruker).
Amino Acid Substitutions of GyrA, GyrB, ParC, and ParE
We estimated amino acid substitutions of DNA gyrase and topoisomerase IV from the nucleotide sequences of gyrA, gyrB, parC, and parE. We compared the substitutions with H. haemolyticus CCUG 12834T or ATCC 33390T (DDBJ/EMBJ/GenBank accession no. LYCK01000011, LYCK01000013, or JTLY01000001), NCTC 10839 (LS483458), M19346 (CP031243), and M28486 (CP031238).
Genomic Analysis
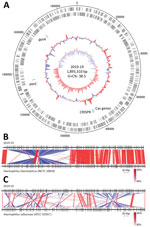
To further characterize the 2019-19 strain in detail, we determined the whole-genome sequence by next generation sequencing (Figure). The genome size was 1,895,310 bp, comprising 1,764 protein-coding sequences, 19 rRNAs, 57 tRNA, and a CRISPR sequence (Figure panel A). Comparing the whole-genome sequence of 2019-19 with that of H. influenzae ATCC 33391T resulted in a dDDH score of 43.4% (95% CI 40.9%–46.0%) and ANIb score of 90.90%, suggesting an extremely low similarity. In contrast, although we observed a large inversion, we found 2019-19 comparatively strongly related to H. haemolyticus NCTC 10839 (Figure, panels B, C). The identity scores for 2019-19 with H. haemolyticus type strain ATCC 33390T were 64.1% (95% CI 61.2%–66.9%) for dDDH and 95.38% for ANIb, suggesting great similarity. To further clarify the genetic classification, we performed phylogenetic analyses using typical species housekeeping genes (16S DNA sequence, adk, pgi, recA, infB) and antimicrobial targeting genes (gyrA, gyrB, parC, parE, and ftsI) (Appendix Figure 1). In all phylogenetic trees using these genes, 2019-19 was classified in the same cluster as H. haemolyticus. In addition, 2019-19 contained a CRISPR sequence but not an IgA protease, which is a putative marker for distinguishing it from H. influenzae (data not shown) (8).
Biochemical Tests
Because we identified this strain as H. influenzae by routine laboratory testing, we also investigated the biochemical characteristics of 2019-19 (Table 1). In comparison with the type strains of both H. haemolyticus and H. influenzae, all biochemical results completely matched with H. influenzae. In addition, we identified species on the basis of these results using Apiweb (bioMérieux), which indicated that 2019-19 had 99.9% identity with H. influenzae. According to the method for H. influenzae described elsewhere, 2019-19 was determined to be biotype II (1).
Mass Spectrometry
MALDI-TOF mass spectrometry analysis is one of the most reliable methods for identifying bacterial species (24,25). The bacterial protein profile was assayed to identify the bacterial species of 2019-19 using this method. In comparison with the database using MALDI Biotyper Compass Explorer version 4.1.60, 2019-19 matched with H. haemolyticus CCUG 12834T with a score 2.21 and was identified as H. haemolyticus. Furthermore, in a phylogenetic dendrogram drawn with representative type strains of Haemophilus spp. (Appendix Figure 2), 2019-19 was located close to the H. haemolyticus type strain ATCC 33390T.
Quinolone Resistance Mechanisms
Antimicrobial susceptibility testing of strain 2019-19 showed high MIC values (16 to ≥64 μg/mL) to levofloxacin, tosufloxacin, and moxifloxacin and susceptibility to antimicrobial agents including penicillins, cephems, and macrolides, but not to quinolones (Table 2). We investigated amino acid substitutions in quinolone-targeting proteins (GyrA, GyrB, ParC, and ParE) by comparing the sequences of H. haemolyticus CCUG 12834T or ATCC 33390T, NCTC10839, M19346, and M28486 (Table 3; Appendix Figure 3). The results revealed that 2019-19 had various amino acid substitutions in GyrA, GyrB, ParC, and ParE, including amino acid substitutions (Ser84Leu, Asp88Tyr in gyrA and Ser84Arg in parC) relevant to reducing susceptibility to quinolones (13,17). We measured the MICs of quinolones in the presence of the efflux pump inhibitors reserpine and PAβN to determine whether the efflux system affected quinolone resistance. There was no substantial difference in the presence or absence of inhibitors. To investigate the origin of this strain, we compared quinolone target genes among Haemophilus spp.; however, we obtained no evidence of recombination (data not shown).
We analyzed high-level quinolone-resistant H. haemolyticus strain 2019-19 isolated from a pediatric patient in an acute care hospital in Japan. The patient had several coexisting diseases and had been under tracheotomy for 2 years but had not been exposed to quinolone for the previous >3 years.
Comparative genome analysis, phylogenetic analysis using typical genes, and MALDI-TOF mass spectrometry analysis indicated that 2019-19 classified into the H. haemolyticus cluster rather than H. influenzae. Absence of IgA protease supported these results (8). In addition, this strain contained CRISPR sequences. Comparing genome sequences in the database, all H. haemolyticus contained CRISPR but H. influenzae sequences did not, which might support that this strain was H. haemolyticus. In contrast, 2019-19 shared biochemical features with H. influenzae. The biotype of 2019-19, biotype II, is the predominant type among H. influenzae and comparatively rare among H. haemolyticus (17,26,27), making it a notable feature of H. haemolyticus 2019-19. Previous studies reported that clinical isolates identified as H. influenzae occasionally included H. haemolyticus without hemolysis (6–8). The biochemical features of 2019-19 likely contributed to this misidentification. Genomic analysis showed 2019-19 contained a large inversion compared with other H. haemolyticus strains; however, the relationship of this inversion with biochemical features was not determined. Bacterial species defined by whole-genome sequence similarity have been reported to be ≈95%–96% ANIb (28) or 70% DDH (29). Although we tentatively identified 2019-19 as H. haemolyticus, these definitions and our ANIb and dDDH values suggest that this strain is a novel subspecies or species. The classification data for Haemophilus spp. are inadequate compared with those of other pathogens and species may need to be reclassified after additional genome and biochemical data are accumulated. Our findings can help improve the accuracy of classification and 2019-19 may be designated a novel subspecies or species in the future (6,30).
H. haemolyticus 2019-19 showed high-level quinolone-resistance and multiple amino acid substitutions in quinolone-targeting proteins, which are known to contribute to high-level quinolone resistance (12–14,17,31). In addition, quinolone-resistant H. parainfluenzae has been reported (32,33) in Taiwan and Europe and these isolates showed various amino acid substitutions in quinolone target genes, like those observed in 2019-19. Frequent use of quinolone can contribute to the emergence of resistant strains, and although this patient had not been exposed to any quinolones during the previous >3 years, she had frequently stayed in medical facilities and other antimicrobial agents had been used to treat her multiple coexisting diseases. Moreover, quinolones have been used for pediatric patients in Japan and the frequency of low-susceptibility strains of H. influenzae has been increasing (9,10,34). There may be selective pressure not only in hospitals but also in communities. In fact, 2019-19 contained a large inversion in the genome and partially differed from H. haemolyticus, suggesting substantial genetic recombination and rearrangement for this strain.
Among the study’s limitations, we analyzed only 1 high-level quinolone-resistant H. haemolyticus and its prevalence in both community and clinical settings remains unclear. In addition, there was no evidence about whether 2019-19 is a causative pathogen or commensal strain. However, the presence of high-level antimicrobial-resistant Haemophilus spp. in children should be noted, because even commensal bacteria can cause lethal infections in immunocompromised hosts.
The reasons why high-level quinolone-resistant Haemophilus spp. had not been isolated from children are unclear. A previous report suggested that quinolones have not been used to treat pediatric infections (11). In addition, because H. influenzae is a commensal nasopharyngeal bacteria for most children (35), quinolone-resistant strains may be outcompeted by other commensal bacteria. The adaptability of quinolone-resistant strains should be further analyzed.
In conclusion, our findings reveal the existence of high-level quinolone-resistant Haemophilus spp. strains in children. Horizontal gene transfer between H. influenzae and H. haemolyticus has been observed (36,37), and high-level quinolone-resistant H. influenzae may also emerge. Therefore, the presence of high-level resistance strains should be considered when quinolones are used to treat children.
Ms. Tanaka is a PhD student at Tokyo University of Pharmacy and Life Sciences. Her research interests are epidemiological trends and the antimicrobial resistance mechanism of Haemophilus species.
Acknowledgment
This work was partially supported by Sasakawa Scientific Research (grant no. 2020-4001) from the Japan Science Society (E.T.) and Nagai Memorial Research Scholarship (no. N-202604) from the Pharmaceutical Society of Japan (E.T.).
References
- Nørskov-Lauritsen N. Classification, identification, and clinical significance of Haemophilus and Aggregatibacter species with host specificity for humans. Clin Microbiol Rev. 2014;27:214–40. DOIPubMedGoogle Scholar
- Adachi Y, Ando M, Morozumi M, Ubukata K, Iwata S. Genotypic characterization of Haemophilus influenzae isolates from paediatric patients in Japan. J Med Microbiol. 2018;67:695–701. DOIPubMedGoogle Scholar
- Kilian M. Genus III. Haemophilus. Winslow, Broadhurst, Buchanan, Rogers and Smith 1917. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s manual of systematic bacteriology, 2nd ed., vol. 2. The proteobacteriaceae. Part B. The gammaproteobacteria. New York: Springer; 2005. p. 883–904.
- Murphy TF, Brauer AL, Sethi S, Kilian M, Cai X, Lesse AJ. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J Infect Dis. 2007;195:81–9. DOIPubMedGoogle Scholar
- Kirkham LA, Wiertsema SP, Mowe EN, Bowman JM, Riley TV, Richmond PC. Nasopharyngeal carriage of Haemophilus haemolyticus in otitis-prone and healthy children. J Clin Microbiol. 2010;48:2557–9. DOIPubMedGoogle Scholar
- McCrea KW, Xie J, LaCross N, Patel M, Mukundan D, Murphy TF, et al. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J Clin Microbiol. 2008;46:406–16. DOIPubMedGoogle Scholar
- Mukundan D, Ecevit Z, Patel M, Marrs CF, Gilsdorf JR. Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J Clin Microbiol. 2007;45:3207–17. DOIPubMedGoogle Scholar
- Hotomi M, Kono M, Togawa A, Arai J, Takei S, Ikeda Y, et al. Haemophilus influenzae and Haemophilus haemolyticus in tonsillar cultures of adults with acute pharyngotonsillitis. Auris Nasus Larynx. 2010;37:594–600. DOIPubMedGoogle Scholar
- Seyama S, Wajima T, Yanagisawa Y, Nakaminami H, Ushio M, Fujii T, et al. Rise in Haemophilus influenzae with reduced quinolone susceptibility and development of a simple screening method. Pediatr Infect Dis J. 2017;36:263–6. DOIPubMedGoogle Scholar
- Tanaka E, Hara N, Wajima T, Ochiai S, Seyama S, Shirai A, et al. Emergence of Haemophilus influenzae with low susceptibility to quinolones and persistence in tosufloxacin treatment. J Glob Antimicrob Resist. 2019;18:104–8. DOIPubMedGoogle Scholar
- Yokota S, Ohkoshi Y, Sato K, Fujii N. Emergence of fluoroquinolone-resistant Haemophilus influenzae strains among elderly patients but not among children. J Clin Microbiol. 2008;46:361–5. DOIPubMedGoogle Scholar
- Cherkaoui A, Gaïa N, Baud D, Leo S, Fischer A, Ruppe E, et al. Molecular characterization of fluoroquinolones, macrolides, and imipenem resistance in Haemophilus influenzae: analysis of the mutations in QRDRs and assessment of the extent of the AcrAB-TolC-mediated resistance. Eur J Clin Microbiol Infect Dis. 2018;37:2201–10. DOIPubMedGoogle Scholar
- Puig C, Tirado-Vélez JM, Calatayud L, Tubau F, Garmendia J, Ardanuy C, et al. Molecular characterization of fluoroquinolone resistance in nontypeable Haemophilus influenzae clinical isolates. Antimicrob Agents Chemother. 2015;59:461–6. DOIPubMedGoogle Scholar
- Tateda K, Ohno A, Ishii Y, Murakami H, Yamaguchi K; Levofloxacin surveillance group. Investigation of the susceptibility trends in Japan to fluoroquinolones and other antimicrobial agents in a nationwide collection of clinical isolates: A longitudinal analysis from 1994 to 2016. J Infect Chemother. 2019;25:594–604. DOIPubMedGoogle Scholar
- Yamada S, Seyama S, Wajima T, Yuzawa Y, Saito M, Tanaka E, et al. β-Lactamase-non-producing ampicillin-resistant Haemophilus influenzae is acquiring multidrug resistance. J Infect Public Health. 2020;13:497–501. DOIPubMedGoogle Scholar
- Ishiwada N, Fujimaki K, Matsumoto T, Kiyota H, Tateda K, Sato J, et al. Nationwide surveillance of bacterial pathogens isolated from children conducted by the surveillance committee of Japanese Society of Chemotherapy, the Japanese Association for Infectious Diseases, and the Japanese Society for Clinical Microbiology in 2017: General overview of pathogenic antimicrobial susceptibility. J Infect Chemother. 2021;27:139–50. DOIPubMedGoogle Scholar
- Marti S, Puig C, de la Campa AG, Tirado-Velez JM, Tubau F, Domenech A, et al. Identification of Haemophilus haemolyticus in clinical samples and characterization of their mechanisms of antimicrobial resistance. J Antimicrob Chemother. 2016;71:80–4. DOIPubMedGoogle Scholar
- Clinical Laboratory and Standards Institute. Performance standards for antimicrobial susceptibility testing (M100). 29th ed. Wayne (PA): The Institute; 2019.
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. DOIPubMedGoogle Scholar
- Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31. DOIPubMedGoogle Scholar
- Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. DOIPubMedGoogle Scholar
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52–7.
- Christensen H, Kuhnert P, Olsen JE, Bisgaard M. Comparative phylogenies of the housekeeping genes atpD, infB and rpoB and the 16S rRNA gene within the Pasteurellaceae. Int J Syst Evol Microbiol. 2004;54:1601–9. DOIPubMedGoogle Scholar
- Bruin JP, Kostrzewa M, van der Ende A, Badoux P, Jansen R, Boers SA, et al. Identification of Haemophilus influenzae and Haemophilus haemolyticus by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2014;33:279–84. DOIPubMedGoogle Scholar
- Saffert RT, Cunningham SA, Ihde SM, Jobe KE, Mandrekar J, Patel R. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J Clin Microbiol. 2011;49:887–92. DOIPubMedGoogle Scholar
- Collins S, Vickers A, Ladhani SN, Flynn S, Platt S, Ramsay ME, et al. Clinical and molecular epidemiology of childhood invasive nontypeable Haemophilus influenzae disease in England and Wales. Pediatr Infect Dis J. 2016;35:e76–84. DOIPubMedGoogle Scholar
- Lulitanond A, Chanawong A, Pienthaweechai K, Sribenjalux P, Tavichakorntrakool R, Wilailuckana C, et al. Prevalence of β-lactamase-negative ampicillin-resistant haemophilus influenzae isolated from patients of a teaching hospital in Thailand. Jpn J Infect Dis. 2012;65:122–5.PubMedGoogle Scholar
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–31. DOIPubMedGoogle Scholar
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. DOIPubMedGoogle Scholar
- Harris TM, Price EP, Sarovich DS, Nørskov-Lauritsen N, Beissbarth J, Chang AB, et al. Comparative genomic analysis identifies X-factor (haemin)-independent Haemophilus haemolyticus: a formal re-classification of ‘Haemophilus intermedius’. Microb Genom. 2020;6:
e000303 . DOIPubMedGoogle Scholar - Georgiou M, Muñoz R, Román F, Cantón R, Gómez-Lus R, Campos J, et al. Ciprofloxacin-resistant Haemophilus influenzae strains possess mutations in analogous positions of GyrA and ParC. Antimicrob Agents Chemother. 1996;40:1741–4. DOIPubMedGoogle Scholar
- Chang CM, Tang HJ, Wang LR, Shih HI, Huang CC, Lee NY, et al. Increasing resistance to fluoroquinolones among Haemophilus species in Southern Taiwan. J Microbiol Immunol Infect. 2017;50:258–60. DOIPubMedGoogle Scholar
- Rodríguez-Martínez JM, López-Hernández I, Pascual A. Molecular characterization of high-level fluoroquinolone resistance in a clinical isolate of Haemophilus parainfluenzae. J Antimicrob Chemother. 2011;66:673–5. DOIPubMedGoogle Scholar
- Mikasa K, Aoki N, Aoki Y, Abe S, Iwata S, Ouchi K, et al. JAID/JSC guidelines for the treatment of respiratory infectious diseases: the Japanese Association for Infectious Diseases/Japanese Society of Chemotherapy—the JAID/JSC guide to clinical management of infectious disease/Guideline-preparing Committee Respiratory Infectious Disease WG. J Infect Chemother. 2016;22(Suppl):S1–65. DOIPubMedGoogle Scholar
- Wouters I, Desmet S, Van Heirstraeten L, Herzog SA, Beutels P, Verhaegen J, et al. NPcarriage Study Group. How nasopharyngeal pneumococcal carriage evolved during and after a PCV13-to-PCV10 vaccination programme switch in Belgium, 2016 to 2018. Euro Surveill. 2020;25:
1900303 . DOIGoogle Scholar - Takahata S, Ida T, Senju N, Sanbongi Y, Miyata A, Maebashi K, et al. Horizontal gene transfer of ftsI, encoding penicillin-binding protein 3, in Haemophilus influenzae. Antimicrob Agents Chemother. 2007;51:1589–95. DOIPubMedGoogle Scholar
- Witherden EA, Bajanca-Lavado MP, Tristram SG, Nunes A. Role of inter-species recombination of the ftsI gene in the dissemination of altered penicillin-binding-protein-3-mediated resistance in Haemophilus influenzae and Haemophilus haemolyticus. J Antimicrob Chemother. 2014;69:1501–9. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleOriginal Publication Date: December 08, 2021
1These authors contributed equally to this article.
Table of Contents – Volume 28, Number 1—January 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Takeaki Wajima, Ph.D, Meijo University, 150 Yagotoyama, Tenpaku, Nagoya 468-8503, Japan
Top