Volume 28, Number 1—January 2022
Dispatch
Intercontinental Spread of Eurasian Highly Pathogenic Avian Influenza A(H5N1) to Senegal
Abstract
In January 2021, Senegal reported the emergence of highly pathogenic avian influenza virus A(H5N1), which was detected on a poultry farm in Thies, Senegal, and in great white pelicans in the Djoudj National Bird Sanctuary. We report evidence of new transcontinental spread of H5N1 from Europe toward Africa.
On December 23, 2020, a single poultry farm composed of 4 barns of laying hens having a total of 102,000 birds in Pout, Thies Region, Senegal, reported increased deaths (mortality rate 58%) to animal health authorities. The clinical signs observed in the affected poultry were edema of the cervical region, cyanosis, congestion of the crests and barbs, and a state of general prostration. Organs and cloacal and oropharyngeal swab specimens collected from dead and sick birds were analyzed at the National Veterinary Laboratory for Livestock and Research (LNERV; Dakar, Senegal), where highly pathogenic avian influenza (HPAI) A (H5N1) virus was confirmed on January 7, 2021.
Later that month, 750 great white pelicans (Pelecanus onocrotalus) (740 juveniles and 10 adults) were found dead by rangers in the Djoudj National Bird Sanctuary, a UNESCO World Heritage site, which is a wetland near the Senegal–Mauritania border. The sanctuary welcomes thousands of Palearctic and Afrotropical migratory birds every year as a refuge, feeding site, and breeding site. On January 15, 2021, the monthly count of birds at Djoudj enacted by the Ministry of Environment documented 8,887 pelicans, for an estimated mortality rate of 8.4% in January 2021. After identifying H5N1 in the dead birds, LNERV analyzed amino acid sequences deduced at the hemagglutinin cleavage site (PLREKRRKR×GLF) on samples from poultry and pelicans, which classified the strain as an HPAI.
Since the emergence of the HPAI H5Nx viruses of the goose/Guangdong (gs/Gd) lineage in 1996, the transcontinental spread of the virus to Africa has been described at least 3 times (1). According to available data, no incursions have involved Senegal before. This unprecedented geographic spread raises questions about the mechanisms of emergence and dissemination of HPAI H5N1 in this country. To determine the origin and transmission pathways of the virus, we analyzed the complete genome of 4 HPAI H5N1 viruses collected in Senegal from domestic and wild birds, studying their spatial diffusion dynamics.
A total of 8 clinical samples were submitted to the World Organisation for Animal Health Reference Laboratory and to the Food and Agriculture Organization of the United Nations Reference Center for Avian Influenza and Newcastle Disease at the Istituto Zooprofilattico Sperimentale delle Venezie (Legnaro, Padova, Italy) for confirmatory diagnosis and genetic characterization of the identified viruses. HPAI H5N1 was identified in all submitted samples by molecular analysis, confirming the results from LNERV. Because of the low viral load, whole-genome sequences were successfully generated from only 4 of 8 samples (Table) collected from poultry and wild birds, as previously described (1).
The phylogenetic analysis of the hemagglutinin gene revealed that the 4 HPAI H5N1 viruses from Senegal belong to clade 2.3.4.4b and cluster not only together but also with the HPAI H5N1 viruses that have been circulating in Europe since October 2020 (99.8%–99.9% nucleotide similarity) (Appendix Figure 1) (2,3). In particular, the HPAI H5N1 viruses from Senegal cluster together in the phylogenetic trees of all 8 gene segments and are closely related to HPAI H5N1 viruses identified in the Netherlands, United Kingdom, and Italy during October–December 2020 (98.8%–100% nucleotide similarity) (Appendix Figures 1–8). These findings suggest virus introduction in the country was likely caused by wild birds’ migration routes from Europe.
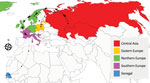
To reconstruct the spatial spread and estimate time of virus introduction into Senegal, we performed a phylogeographic analysis of the hemagglutinin gene in BEAST version 1.10.4 (4). We defined 5 discrete geographic regions: Central Asia, Northern Europe, Eastern Europe, Southern Europe, and Senegal. The mean time to the most recent common ancestor of the HPAI H5N1 viruses from Senegal was estimated to be November 2020 (95% HPD interval October–December 2020). The genetic spatial analysis indicated that the virus had spread from Southern Europe to Senegal, which suggests that West Africa likely acted as the ecologic sink of the HPAI H5N1 viruses circulating in Europe (Figure; Appendix Figure 9). Because availability of viral sequences from different countries could affect phylogeographic analyses, having a large number of sequences available is vital to obtain accurate and reliable results.
These evolutionary and spatial investigations indicate that the H5N1 outbreaks in Senegal did not emerge from local evolution of H5N1 viruses in Africa. These new viruses seem to have been introduced in fall 2020 from Eurasia through migratory birds flying southwest for winter. The estimated time to the most recent common ancestor (October–December 2020) and the long branches that separate the Senegal viruses from progenitors in Europe suggest an undetected virus circulating in the area, likely in wild birds.
The Djoudj National Bird Sanctuary, located in the Senegal River delta along the East Atlantic Flyway, is a sanctuary for large breeding waterbirds, including great white pelicans. H5N1 caused the death of hundreds of pelicans there. Before the emergence of the HPAI H5Nx viruses of the Gs/GD lineage, infection with avian influenza virus of pelicans was rarely reported (5). The incursion of the Gs/GD lineage has resulted in numerous fatal infections in this species. On the basis of data from the avian influenza passive surveillance system implemented in Europe during 2005–2017, an HPAI detection rate of 9.5% has been estimated in great white pelicans (6). This species is highly gregarious, behavior that could have promoted the spread of HPAI in these birds in Senegal. Unfortunately, surveillance of wild and domestic birds near where the H5N1-infected pelicans were identified did not shed light on the species responsible for introducing the virus. The wetlands of Senegal are inhabited by millions of aquatic bird species, including Garganey (Anas querquedula), Northern pintail (Anas acuta), Northern shoveler (Spatula clypeata), Eurasian teal (Anas crecca), Eurasian wigeon (Mareca penelope), Common pochard (Aythya farina), and Tufted duck (Aythya fuligula), many of which have had a role in the spread of Gs/GD-lineage H5 viruses (7,8). This bird population has been hardly affected by the 2020–2021 epidemic in Europe, during which HPAI H5 virus infections were reported in apparently healthy birds (2). Therefore, even considering that most of the great white pelicans residing in the Djoudj National Bird Sanctuary are deemed to be sedentary (9), species other than pelicans might have been involved in introducing and spreading H5N1 within Senegal or to neighboring countries reporting HPAI H5N1, including Niger, Nigeria, Mauritania, and Mali.
Recent reports of H5N1 in Senegal, Mauritania, and Mali indicate an unprecedented westward spread of the virus in Africa (10). However, the lack of genetic information on the viruses detected in these countries makes it difficult to reconstruct the exact number of virus introductions and dynamics of virus dissemination, and both poultry trade and wild bird movements remain valid candidate pathways. Moreover, after the outbreaks of HPAI in poultry, 2 states in Nigeria reported 7 suspected human cases of avian influenza H5N1, 4 in Kano and 3 in Plateau. These cases confirm the importance of One Health joint activities by public human and animal health sectors to contain and monitor virus spread and the emergence of novel viruses of major concern (11).
There is still much to learn about the ecology of these viruses in the wild bird population; detection of the 2.3.4.4b clade in Senegal demonstrates that predicting the dissemination trajectories of these viruses is difficult. No system yet exists that can prevent the virus from following wild bird movements. Efforts are needed to regulate poultry movements and develop risk-based surveillance in wild birds in Africa to detect newly introduced and circulating viruses, reduce the likely spread to poultry, and limit the risk for exposure of humans to infected birds.
Dr. Lo is a microbiologist and head of the Laboratory of Bacteriology and Avian Pathology at the National Laboratory of Livestock and Veterinary Research at Institut Sénégalais de Recherches Agricoles, Dakar-Hann, Senegal. Her research interests include epidemiologic surveillance of cross-border diseases such as avian influenza and serologic and molecular epidemiology, gene flow, and the One Health approach.
Acknowledgments
LNERV extends its sincere appreciation to support from the International Atomic Energy Agency (IAEA) through the sequencing services of the Animal Production and Health Section of the Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture for the partial sequencing of the hemagglutinin gene, which made it possible to confirm the HPAI H5N1 subtype. We also thank all the operators engaged in field sampling who made the investigation possible: the field veterinary services in Thies and St Louis regions and the regional and local administrations in these regions for their technical and administrative support. We would also like to thank the FAO’s Emergency Centre for Transboundary Animal Diseases Senegal for funding the field investigation. Finally, we thank the authors and the originating and submitting laboratories of the sequences from the GISAID EpiFlu Database on which this research is partly based (Appendix Table).
Support for this work was provided by UN-FAO with funding from US Agency for International Development under the OSRO/GLO/501/USA, OSRO/GLO/506/USA and OSRO/GLO/507/USA, TCP/SFC/3704 and OSRO/EGY/501/USA projects (PO 345340). The content of this article is the responsibility of the author(s) and does not necessarily reflect the views of UN-FAO, USAID, or the United States Government. The research leading to these results was also been partly funded by the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 725422-ReservoirDOCS).
This article is in memory of our beloved colleague Matteo Griggio, a lively behavioral ecologist strongly committed to nature conservation, who prematurely passed away on May 14, 2020.
References
- Fusaro A, Zecchin B, Vrancken B, Abolnik C, Ademun R, Alassane A, et al. Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza. Nat Commun. 2019;10:5310. DOIPubMedGoogle Scholar
- Adlhoch C, Fusaro A, Gonzales JL, Kuiken T, Marangon S, Niqueux É, et al.; European Food Safety Authority, European Centre for Disease Prevention and Control and European Union Reference Laboratory for Avian Influenza. Avian influenza overview August - December 2020. EFSA J. 2020;18:
e06379 .PubMedGoogle Scholar - Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec. 2018;93:142–52.PubMedGoogle Scholar
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5:
e1000520 . DOIPubMedGoogle Scholar - Stallknecht DE, Shane SM. Host range of avian influenza virus in free-living birds. Vet Res Commun. 1988;12:125–41. DOIPubMedGoogle Scholar
- European Food Safety Authority. Target list of wild bird species for passive surveillance of H5 HPAI viruses in the EU, based on passive surveillance data from 2005 to 2017 [cited 2021 Oct 7]. https://www.izsvenezie.com/documents/reference-laboratories/avian-influenza/useful-resources/wild-bird-target-species-for-passive-surveillance.pdf
- Global Consortium for H5N8 and Related Influenza Viruses. Role for migratory wild birds in the global spread of avian influenza H5N8. Science. 2016;354:213–7. DOIGoogle Scholar
- Gilbert M, Xiao X, Domenech J, Lubroth J, Martin V, Slingenbergh J. Anatidae migration in the western Palearctic and spread of highly pathogenic avian influenza H5NI virus. Emerg Infect Dis. 2006;12:1650–6. DOIPubMedGoogle Scholar
- International Union for Conservation of Nature and Natural Resources. Pelecanus onocrotalus (Great White Pelican). The IUCN red list of threatened species 2018 [cited 2021 Oct 7]. https://www.iucnredlist.org/species/22697590/132595920
- Food and Agriculture Organization of the United Nations. Sub-Saharan Africa HPAI situation update [cited 2021 May 24]. https://www.fao.org/ag/againfo/programmes/en/empres/HPAI_Africa/situation_update.html
- ProMED-mail. PRO/AH/EDR> Avian influenza, human (05): Nigeria, A/H5 [cited 2021 May 24]. https://promedmail.org, archive no. 20210407.8294492.
Figure
Table
Cite This ArticleOriginal Publication Date: December 13, 2021
1These first authors contributed equally to this article.
Table of Contents – Volume 28, Number 1—January 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Bianca Zecchin, World Organisation for Animal Health Reference Laboratory, Food and Agriculture Organization of the United Nations Reference Center for Animal Influenza and Newcastle disease, Istituto Zooprofilattico Sperimentale delle Venezie, viale dell’università 10, 35020 Legnaro, Padova, Italy
Top