Volume 28, Number 1—January 2022
Dispatch
Multistate Outbreak of Salmonella enterica Serovar Heidelberg with Unidentified Source, Australia, 2018–2019
Abstract
We report a multistate Salmonella enterica serovar Heidelberg outbreak in Australia during 2018–2019. Laboratory investigation of cases reported across 5 jurisdictions over a 7-month period could not identify a source of infection but detected indicators of severity and invasiveness. The hospitalization rate of 36% suggested a moderately severe clinical picture.
Salmonella enterica serovar Heidelberg is a frequently identified serotype among infections in humans in North America, East Africa, and Asia but is uncommon in Australia. An average of 37 cases of Salmonella Heidelberg were notified in Australia annually in 2009–2017, predominantly overseas acquired (1). Six outbreaks have been reported nationally since 1995; 1 outbreak in 1996 had >500 cases, but most have <7 cases (R. Bell, pers. comm. [email], 2020 Jun 16). We report a national outbreak of Salmonella Heidelberg infection across 5 jurisdictions over 7 months.
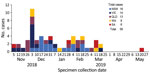
In December 2018, OzFoodNet, Australia’s government-based network for enhanced foodborne disease surveillance, noted that Salmonella Heidelberg cases diagnosed in November (15 cases) were above the national historical 5-year mean (2.4 cases). New South Wales (NSW) and Victoria initiated separate investigations during December 2018–February 2019; neither developed a hypothesis regarding potential sources of infection. In February–March 2019, whole-genome sequencing (WGS) analysis of available isolates identified 36 highly related cases, 12 each from Queensland, NSW, and Victoria. Concurrent Salmonella Heidelberg infections with WGS pending were subsequently identified in other states: South Australia (SA) (N = 4) and Western Australia (WA) (N = 3). Queensland cases were not initially investigated because Salmonella Heidelberg is more common in this state; Queensland contributed 43% of cases in Australia during 2009–2017 (1). Outbreak cases were reported across multiple jurisdictions peaking in early December 2018 and continuing through late March 2019, with an outlying case reported in May 2019 (Figure). After confirmation of phylogenetic relatedness and previous jurisdictional inability to identify a common source, OzFoodNet commenced a multijurisdictional outbreak investigation in March 2019. However, case numbers declined soon after, preventing more rigorous, prospective epidemiologic investigation.
We identified 59 outbreak cases in 5 jurisdictions (58 laboratory-confirmed, 1 epidemiologically-linked): NSW (18/59, 31%), Victoria (14/59, 24%), Queensland (13/59, 22%), WA (8/59, 14%), and SA (6/59, 10%) (Table). Case-patients were 2 months–95 (median 43) years of age. None had a history of international travel; 3 case-patients reported interstate travel during their exposure period.
Thirty-nine case-patients completed interviews using telephone-administered hypothesis-generating questionnaires. The remaining 20 patients were not investigated because they refused interviews, they could not be contacted, or their diagnosis dates precluded obtaining a reliable food history. Binomial probability analysis compared case-patient food exposures to background rates estimated from a Victoria population survey, accounting for seasonality, from November 2014–October 2016 (2).
Probability calculations highlighted potential foods of interest, including cooked chicken (p<0.001), macadamia nuts (p = 0.001), frozen vegetable products (p = 0.005), and lamb (p = 0.005). Global epidemiology suggests Salmonella Heidelberg outbreaks are most likely associated with poultry or eggs (3–5), yet case reporting of poultry products consumed and place of purchase did not identify a common source. Raw macadamia nuts were considered because of their popularity during the Christmas period, along with previous detections of Salmonella Heidelberg in Queensland (6). Because epidemiologic evidence was insufficient to develop a strong hypothesis for any single food item, sampling was not considered feasible; decreasing case numbers precluded an analytic study.
We consolidated sequence data, including isolates sequenced in jurisdictional laboratories. We assessed genetic relatedness among isolates of Salmonella Heidelberg cases at the Queensland Health Forensic and Scientific Services laboratory by generating core genome multilocus sequence typing (cgMLST) complex types (<7 allele differences) using Ridom SeqSphere+ version 5.1.0 (https://www.ridom.de) based on the EnteroBase Salmonella enterica scheme version 2.0 (https://enterobase.warwick.ac.uk). We determined single-nucleotide polymorphism (SNP) differences using Snippy package version 4.3.6 (https://github.com/tseemann/snippy) using Salmonella Heidelberg SL476 (GenBank accession no. NC_011083.1) as a reference. We conducted in silico WGS analysis for antimicrobial-resistance genes by the abricate program version 0.8.10 (https://github.com/tseemann/abricate), using the Resfinder database October 18, 2018, version (https://cge.cbs.dtu.dk/services/ResFinder) to further characterize strains. We investigated the presence of the saf operon using Ridom SeqSphere+ against the saf operon sequence from GenBank (accession no NZ_LS483494). We generated sequences for isolates on the Illumina NextSeq genome sequencing platform (https://www.illumina.com) using the Nextera XT library preparation kit; the WGS read files were deposited in the US National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov) Sequence Read Archive (BioProject ID PRJNA663036 [NSW] and PRJNA660800 [Queensland, Victoria, SA, WA]). We developed our hypothesis using a range of local and international sequence data sources to assess genetic relatedness to outbreak cases; we used 78 human sequences from 5 jurisdictional laboratories, 8 nonhuman Salmonella Heidelberg isolates from a Queensland culture collection from food and animals (macadamia nuts and poultry, caprine, porcine, bovine, equine, and reptile sources), and 86 international sequences downloaded from EnteroBase and the National Center for Biotechnology Information.
The 58 Salmonella Heidelberg isolates available from this outbreak were all multilocus sequence type (ST) 15 and belonged to the same cgMLST complex type (2561); isolates differed within the complex type by 0–3 SNPs/cgMLST alleles, whereas the nearest nonoutbreak genomes had >34 SNP and >16 SNP allele differences (Appendix Figure). This complex type was not identified among a range of international sequences or Australia nonhuman or historical human isolates chosen to inform possible outbreak sources; thus, we could not develop our hypothesis using WGS. No compelling microbiological evidence supported hypotheses of a nut, poultry, or other specific source.
All Australia isolates and 91% of international isolates harbored the antimicrobial-resistance gene fosA7 for fosfomycin (7). Phenotypic analysis of 9 isolates revealed the outbreak strains to be susceptible to antimicrobial drugs including cephalosporins, fluoroquinolones, aminoglycosides, broad-spectrum penicillins, and trimethoprim/sulfamethoxazole.
Of 45 case-patients for whom data were available, 16 (36%) were hospitalized with a median duration of 4 (range 1–18) days, reported by 13 case-patients. Although direct comparison is difficult because of potential confounding by age, the hospitalization rate of 36% was high compared with the rate of 11.6% among 149 US outbreaks of Salmonella Heidelberg from 1973–1997 (3). Hospitalization rates in that outbreak were also higher than that for Salmonella enterica outbreaks in Australia with similar age distributions during 2001–2016 (8).
Although this investigation was unable to capture invasiveness of the outbreak strain, Salmonella Heidelberg has frequently been associated with greater risk for invasive disease than have other commonly reported nontyphoidal Salmonella serotypes, including Typhimurium and Enteritidis (9–14). In the United States, Salmonella Heidelberg is among the 4 most common serotypes isolated from blood; 12%–13% of Salmonella Heidelberg infections resulted in invasive disease in North America, higher than the US Salmonella average of 7% (10–12). A study of invasive nontyphoidal Salmonella infection in Australia similarly found that almost 10% of Salmonella Heidelberg gastrointestinal infections during 2007–2016 were invasive disease, ≈5× higher than Salmonella Typhimurium infections (15). Concern has grown regarding the virulence of a US bovine-related Salmonella Heidelberg outbreak; recent genomic analyses indicated that most identified Salmonella virulence genes are present in most Salmonella Heidelberg strains. These studies highlighted potential contributions of saf fimbrial genes to increased severity via their role in bacterial aggregation, colonization, and biofilm formation (9). The saf operon has been reported generally absent from the Salmonella Heidelberg serovar but was present in a previous outbreak associated with increased severity (9). The saf operon was present in all Australia isolates in this study (Appendix Figure).
We report a national outbreak investigation of a locally uncommon S. enterica serovar of unknown origins in Australia. Although Salmonella Heidelberg outbreaks are relatively uncommon in Australia, given this outbreak’s comparatively high hospitalization rate and the presence of saf fimbrial genes in the implicated strain, future cases warrant prompt investigation to assess severity and invasiveness. A platform for real-time exchange of sequence data in Australia and use of routine WGS for salmonellosis cases, including comparison with local and international strain data, may enable more timely detection of outbreaks.
Ms. Kerr is a Master of Applied Epidemiology Scholar (Australia’s only Field Epidemiology Training Program, Australian National University) and ASEAN-Australia Health Security Fellow. Her research interests focus on infectious disease and public health.
Acknowledgments
We thank all referring pathology laboratories for isolation and referral of Salmonella isolates and the Salmonella reference sections in Public Health Reference laboratories for serotyping of Salmonella isolates. We would also like to thank the OzFoodNet Network, which is funded by the Australian Government Department of Health.
Ethics approval for this work was obtained from the Australian National University Human Research Ethics Committee (protocol 2017/909).
While doing this work, E.K. was supported by the Australian National University Master of Philosophy (Applied Epidemiology) Scholarship, which was funded by Queensland Health.
References
- Department of Health. Salmonella National Notifiable Disease Surveillance System notifications in Australia 2009–2017. Canberra, ACT: Commonwealth of Australia; 2019 [updated 2018 Jul 31; cited 2020 May 6]. http://www9.health.gov.au/cda/source/pub_salmo.cfm
- St. George SC. Applied epidemiology in communicable diseases, Victoria, 2016–2017. Canberra, ACT: Australian National University; 2018 [updated 2020 May 19; cited 2020 Jul 9]. https://openresearch-repository.anu.edu.au/handle/1885/154282
- Chittick P, Sulka A, Tauxe RV, Fry AM. A summary of national reports of foodborne outbreaks of Salmonella Heidelberg infections in the United States: clues for disease prevention. J Food Prot. 2006;69:1150–3. DOIPubMedGoogle Scholar
- Chai SJ, Cole D, Nisler A, Mahon BE. Poultry: the most common food in outbreaks with known pathogens, United States, 1998-2012. Epidemiol Infect. 2017;145:316–25. DOIPubMedGoogle Scholar
- Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, Rosen H, et al.; Salmonella Heidelberg Investigation Team. Salmonella Heidelberg Investigation Team. National outbreak of multidrug-resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One. 2016;11:
e0162369 . DOIPubMedGoogle Scholar - Munck N, Smith J, Bates J, Glass K, Hald T, Kirk MD. Source Attribution of Salmonella in macadamia nuts to animal and environmental reservoirs in Queensland, Australia. Foodborne Pathog Dis. 2020;17:357–64. DOIPubMedGoogle Scholar
- Rehman MA, Yin X, Persaud-Lachhman MG, Diarra MS. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob Agents Chemother. 2017;61:e00410–7. DOIPubMedGoogle Scholar
- Ford L, Moffatt CRM, Fearnley E, Miller M, Gregory J, Sloan-Gardner TS, et al. The epidemiology of Salmonella enterica outbreaks in Australia, 2001–2016. Front Sustain Food Syst. 2018;2:1–8. DOIGoogle Scholar
- Antony L, Behr M, Sockett D, Miskimins D, Aulik N, Christopher-Hennings J, et al. Genome divergence and increased virulence of outbreak associated Salmonella enterica subspecies enterica serovar Heidelberg. Gut Pathog. 2018;10:53. DOIPubMedGoogle Scholar
- Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, et al.; Emerging Infections Program NARMS Working Group. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother. 2011;55:1148–54. DOIPubMedGoogle Scholar
- Otto SJ, Carson CA, Finley RL, Thomas MK, Reid-Smith RJ, McEwen SA. Estimating the number of human cases of ceftiofur-resistant Salmonella enterica serovar Heidelberg in Québec and Ontario, Canada. Clin Infect Dis. 2014;59:1281–90. DOIPubMedGoogle Scholar
- Dutil L, Irwin R, Finley R, Ng LK, Avery B, Boerlin P, et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg Infect Dis. 2010;16:48–54. DOIPubMedGoogle Scholar
- Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D’Angelo M, Hurd S, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. 2008;198:109–14. DOIPubMedGoogle Scholar
- Williamson DA, Lane CR, Easton M, Valcanis M, Strachan J, Veitch MG, et al. Increasing antimicrobial resistance in nontyphoidal Salmonella isolates in Australia from 1979 to 2015. Antimicrob Agents Chemother. 2018;62:e02012–7. DOIPubMedGoogle Scholar
- Parisi A, Crump JA, Stafford R, Glass K, Howden BP, Kirk MD. Increasing incidence of invasive nontyphoidal Salmonella infections in Queensland, Australia, 2007-2016. PLoS Negl Trop Dis. 2019;13:
e0007187 . DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: December 13, 2021
Table of Contents – Volume 28, Number 1—January 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Elenor Kerr, National Centre for Epidemiology and Population Health, Australian National University, Bldg 62 Mills Rd, Canberra, ACT 2601, Australia
Top