Volume 28, Number 2—February 2022
Research Letter
Novel Anaplasmataceae agents Candidatus Ehrlichia hydrochoerus and Anaplasma spp. Infecting Capybaras, Brazil
Figure
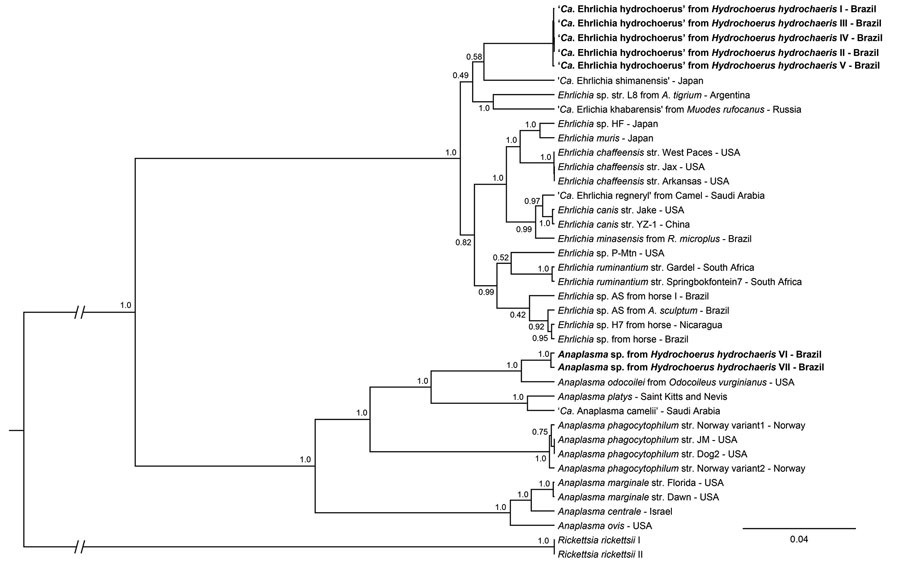
Figure. Phylogenetic analysis of 16S rRNA, sodB, and groEL partial sequences of Candidatus Ehrlichia hydrochoerus and Anaplasma spp. obtained from capybaras (Hydrochoerus hydrochaeris), southern Brazil. These sequences (in bold) and those of other Ehrlichia and Anaplasma species were aligned using MAFFT 7.110 (https://mafft.cbrc.jp/alignment/server). Phylogenetic analyses of each gene were based on Bayesian inference using Beast version 1.8.4 (https://beast.community/index.html). We performed 3 independent runs of 100 million generations of Monte Carlo Markov chain with 1 sampling/10,000 generations and a 10% burn-in. We estimated substitution models as generalized time reversible plus gamma for 16S rRNA (A), Hasegawa–Kishino–Yano plus gamma for sodB (B), and Tamura–Nei plus gamma for groEL (C) genes on the basis of Akaike information criterion by using jModeltest version 2.1.10 (https://github.com/ddarriba/jmodeltest2/releases/tag/v2.1.10r20160303). The tree was rooted with Rickettsia rickettsii (GenBank accession nos. CP000766.3 and CP018913.1). Complete GenBank accession numbers are listed in the Appendix . Scale bar indicates number of substitutions per site. Ca., Candidatus.