Volume 28, Number 2—February 2022
Research
Widespread Detection of Multiple Strains of Crimean-Congo Hemorrhagic Fever Virus in Ticks, Spain
Figure 2
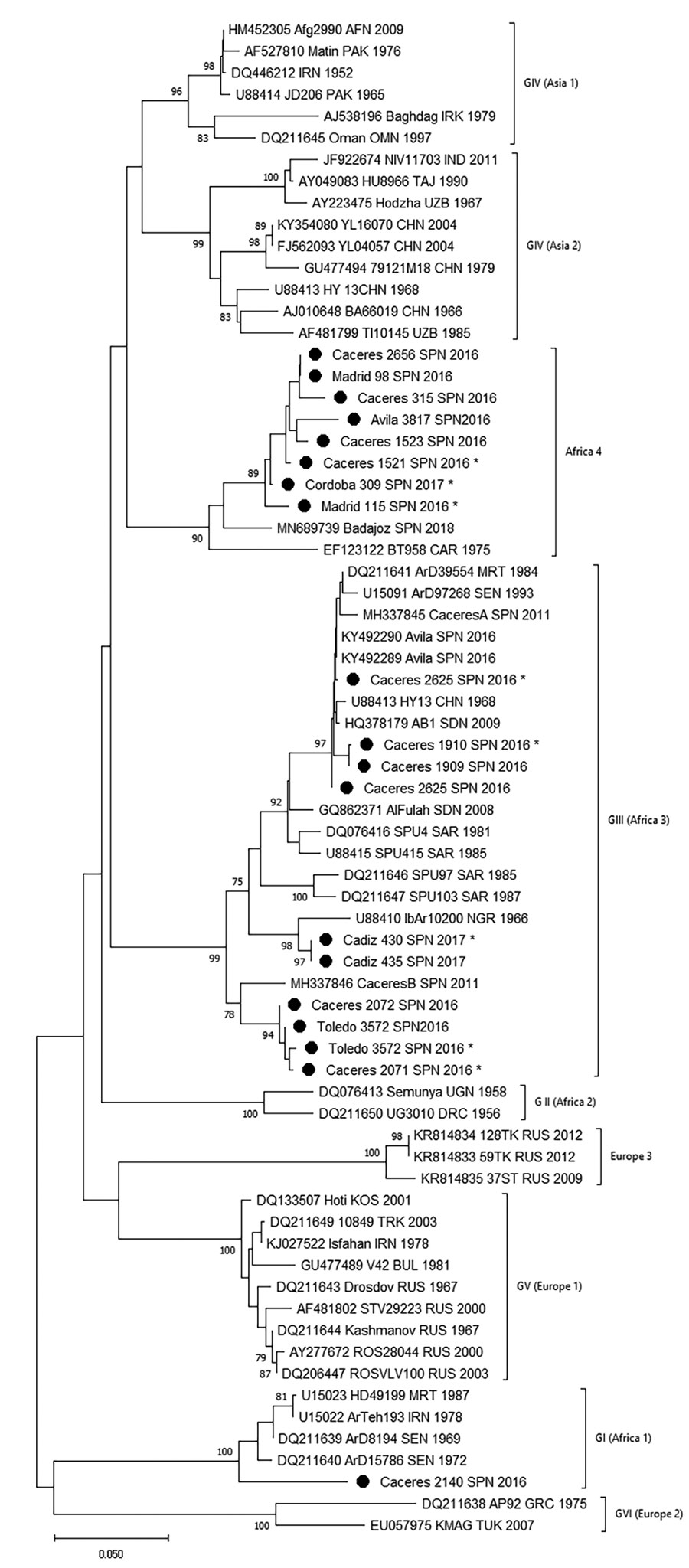
Figure 2. Phylogenetic tree obtained for strains of Crimean-Congo hemorrhagic fever virus detected in Spain (black dots) and other sequences downloaded from GenBank. We built the phylogenetic tree using the neighbor-joining method based on partial (175 nt) sequences of the virus small segment. Numbers in nodes indicate bootstrap values for the groups; values <75 are not shown. Strains detected from Spain are named by geographic origin, locality sampling site, and sampling year; other sequences are named by GenBank accession number, strain, geographic origin, and sampling year. Asterisks indicate sequences from this study that have been submitted to the EMBL (https://www.embl.org) and GenBank databases. Genotypes are indicated by Roman numerals: I, West Africa (Africa 1); II, Central Africa (Africa 2); III, South and West Africa (Africa 3); IV, Middle East/Asia, divided into groups corresponding to groups Asia 1 and Asia 2; V, Europe/Turkey (Europe 1); VI, Greece (Europe 2). Using guidelines published elsewhere (25,26), we then named and labeled the genotypes with equivalent clade nomenclature indicated in parentheses. Scale bar indicates substitutions/site (evolutionary distance).