Volume 28, Number 3—March 2022
Research
Genomic and Phenotypic Insights for Toxigenic Clinical Vibrio cholerae O141
Figure 3
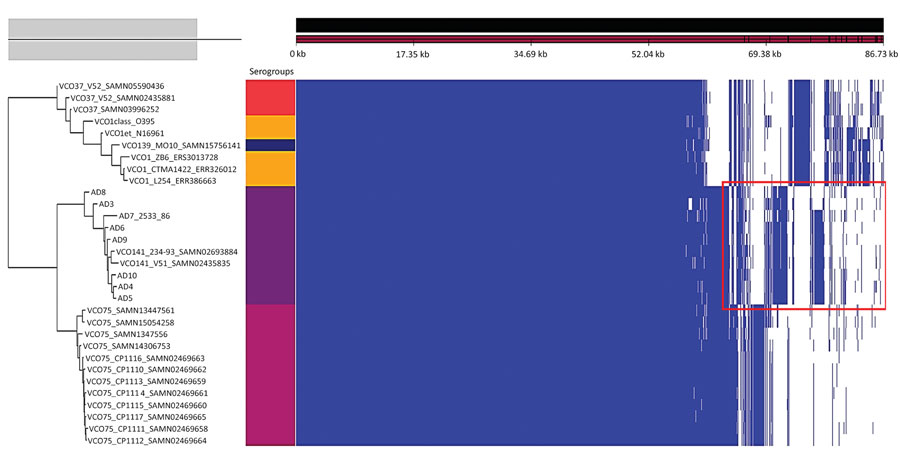
Figure 3. Gene presence/absence map of the pangenome of 31 isolates from cholera toxin‒positive Vibrio cholerae serogroups. The red rectangle in the accessory genome indicates the conserved unique coding sequences that are specific to the serogroup O141. Each serogroup is associated with a color block: O37 (red), O1 (orange), O139 (dark blue), O141 (purple), O75 (Marron). Scale bar indicates nucleotide substitutions per site.
Page created: January 05, 2022
Page updated: February 21, 2022
Page reviewed: February 21, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.