Volume 28, Number 6—June 2022
Dispatch
Identification of Human Case of Avian Influenza A(H5N1) Infection, India
Abstract
A 11-year-old boy with acute myeloid leukemia was brought for treatment of severe acute respiratory infection in the National Capital Region, New Delhi, India. Avian influenza A(H5N1) infection was laboratory confirmed. Complete genome analysis indicated hemagglutinin gene clade 2.3.2.1a. We found the strain to be susceptible to amantadine and neuraminidase inhibitors.
Avian influenza viruses remain major threats worldwide, responsible for multiple outbreaks among poultry and episodes of transmission to humans. During January 2003–February 3, 2022, there were 862 reported cases of human infection with avian influenza A(H5N1) virus in 18 countries, resulting in a 53% case-fatality rate (https://www.who.int/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai-20220401.pdf).
The first outbreak of highly pathogenic avian influenza H5N1 in poultry in India, which occurred in January 2006 in Maharashtra, was caused by clade 2.2 (1); subsequent yearly outbreaks reported in poultry across the country were caused by newer clades 2.3.2.1 and 2.3.2.1c (2–4). Avian influenza surveillance in poultry revealed the presence of low-pathogenicity H9N2 and H4N6 viruses (5). On March 15, 2019, a human case of low-pathogenicity avian influenza A(H9N2) was detected in India (6). To date, 371 H5N1 and H5N8 avian influenza outbreaks in domestic or wild birds have been recorded in 15 of 28 states in India (https://empres-i.apps.fao.org/diseases). The first outbreaks of highly pathogenic avian influenza H5N8 in Europe were reported in August 2020 and since have been reported in poultry and wild birds in several countries in Europe, Asia, and Africa (https://web.oie.int/downld/SG/2021/A_88SG_2.pdf).
An 11-year old boy who had acute myeloid leukemia diagnosed in June 2021 in the department of pediatrics at the All India Institute of Medical Sciences (AIIMS) in New Delhi was brought in for treatment of fever, cough, coryza, and breathing difficulty in the first week of July 2021. The patient was a resident of Gurugram, National Capital Region (Delhi), India. His clinical work-up showed febrile neutropenia with pneumonia and shock, which progressed to acute respiratory distress syndrome, so he was mechanically ventilated. He then developed multiorgan dysfunction, which ultimately resulted in his death on July 12, 2021. An in-depth interview with family members indicated that the patient often frequented a family-owned poultry business and may have been exposed to birds with undetected infection, although no infected domestic or wild avian sources or any environmental contamination had been reported in or around the residence of the child.
Staff in The AIIMS department of pediatrics referred nasopharyngeal (NP) swab specimens collected on July 7 and bronchoalveolar lavage (BAL) fluid collected on July 11 to the department of microbiology for respiratory virus testing, which used a Fast Track Diagnostics Respiratory Pathogens 21 kit and real-time PCR for influenza (https://www.siemens-healthineers.com) for diagnosis. Both BAL and NP samples tested positive for influenza A and influenza B Victoria lineage. Influenza A type could not be determined, so we referred samples to the National Influenza Centre at the Indian Council of Medical Research of the National Institute of Virology (Pune, India) for differential influenza diagnosis. The real-time reverse transcription PCR for avian influenza viruses H5Nx, H7N9, H9N2, and H10N8 was performed as documented elsewhere (7,8); results were positive for an H5Nx virus (cycle threshold value for H5 was 25). To confirm the subtype A/H5 identity, short fragments of multiple genes were sequenced: 230-bp matrix, 400-bp hemagglutinin (HA), 600-bp neuraminidase (NA), 600-bp nonstructural), and the influenza B HA gene, and analyzed results were analyzed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). We isolated and identified strain A/India/NIV-SARI-4571/2021 (H5N1) at a Biosafety Level 4 laboratory using MDCK cells.
We then generated whole-genome sequences from the original clinical NP samples and BAL-and MDCK-grown for passages 1 and 2 isolates using the Miniseq NGS Platform (Illumina, https://www.illumina.com) and a de-novo assembly program using QIAGEN CLC Genomics software 10.1.1 (https://www.quiagen.com). We constructed phylogenetic trees for 8 genes of A/India/NIV-SARI-4571/2021 (H5N1) virus using the neighbor-joining model with a Tamura-Nei nucleotide substitution performing 1,000 replicates of bootstrap support implemented in MEGA 7 (https://megasoftware.net) software. We submitted sequences to GenBank (accession nos. OL311384–91).
The sequences generated for the original clinical sample and the passaged virus were identical, suggesting no passage-induced mutations. We performed BLAST analysis of all 8 genes of A/India/NIV-SARI-4571/2021. The HA gene showed 97% identity with A/duck/Bangladesh/32003/2017 (H5N1). The polymerase basic (PB) 1 and nonstructural genes showed 97% nucleotide similarity to the avian influenza A/duck/Mongolia/729/2019 (H4N6) virus, suggesting probable reassortment.
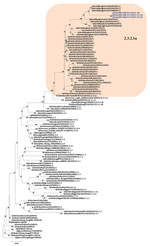
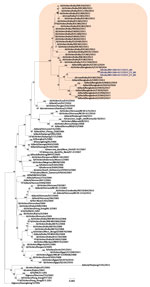
Phylogenetic analysis of the HA gene demonstrated that the virus belonged to clade 2.3.2.1a (Figure 1) and clustered with the A/duck/Bangladesh 2017 (H5N1)–like strain. Clade 2.3.2.1a has a H9N2-like PB1 gene and is the dominant clade in poultry in many countries, including Vietnam (9), India (3), Bangladesh, Bhutan, and Nepal (10). The NA gene clustered with an A/crow/India/01CA02/2014 (H5N1)–like strain (Figure 2). Phylogenetic trees indicate locations for the other 6 genes of the study strain (Appendix Figure 1).
Using the WHO/CDC H5N1 inventory (11), we reviewed potential markers for the newly identified A/India/NIV-SARI-4571/2021 strain. The HA protein (H5) possessed a multiple basic amino acid cleavage site motif (PQRERRRKR*G), resulting in a highly pathogenic strain of clade 2.3.2.1 viruses. The sequence of the 220-loop receptor-binding site between amino acids Q222 and G224 remains conserved for the avian α 2–3 receptor.
We did not observe the NA and matrix 2 gene mutations responsible for neuraminidase inhibitors and amantadine resistance in the study strain. The virus remains purely avian-adapted, and we observed no markers for adaptation in mammals or pathogenicity for humans (Appendix Figure 2). We also did not observe PB2 hallmark mutations E627K and D701N, responsible for host adaptation and virulence in mammals in the study strain (12). The PDZ ligand domain (ESEV) at the C terminus remained conserved. We observed further compensatory mutations during the adaptation of H5N1 in mice, L89V, G309D, T339K, R477G, and I495V of PB2 (13) in the study strain. However, further studies are required to understand their role.
No evidence of H5 antibodies was detected in 18 close contacts of the deceased child. Available information and initial field investigations revealed that no additional cases were detected, indicating low human-to-human transmission. However, unreported high-pathogenicity avian influenza virus continues to exist in traded poultry in India, constituting a substantial risk for further human exposure (https://www.who.int/emergencies/disease-outbreak-news/item/human-infection-with-avian-influenza-a(h5n1)-%EF%BD%B0-india). Although widespread avian influenza outbreaks have been documented globally, only a limited number have shown transmission of avian influenza viruses to humans (14). The severe immunocompromised status of the child in this study may have made him vulnerable, and direct exposure to infected poultry might have been the source of infection. Influenza B virus, simultaneously detected with influenza A in this case-patient, is known to persist in such cases and might have been identified because of a prior infection (15).
In December 2020 and early 2021, outbreaks of avian influenza H5N1 and H5N8 were reported in poultry in 15 states in India; the National Capital Region, Maharashtra, Punjab, and Kerala, in particular, were severely affected. The whole-genome sequencing of A/India/NIV-SARI-4571/2021 (H5N1) virus in our study provides valuable insight into the absence of hallmark mutations responsible for adaptation and virulence in mammals. The strain remained sensitive to amantadine and neuraminidase inhibitors. However, identification of a human H5N1 case in India highlights the need for systemic surveillance at the human–animal interface level.
Dr. Potdar is senior scientist heading the National Influenza Centre at the National Institute of Virology, Pune, India. Her primary research interest is molecular characterization of influenza, SARS-CoV-2, and other respiratory viruses.
Acknowledgments
The authors acknowledge the National Centre for Disease Control, Delhi, for field investigation and Chakradhar Tosh from the Indian Council of Agricultural Research–National Institute of High Security Animal Diseases for sharing the serum samples of close contacts of the case. The authors from the National Institute of Virology, Pune, acknowledge Shailesh Pawar for sharing the serological protocols, the National Influenza Centre team for diagnosis, and the biosafety level 4 laboratory for sequencing and virus isolation and serology testing. The authors from AIIMS acknowledge the Department of Health Research and the Indian Council of Medical Research, Government of India, for supporting the virology laboratory as a regional laboratory under the Virus Research and Diagnostic Laboratory network.
Financial support was provided by the Indian Council of Medical Research, Department of Health Research, New Delhi.
References
- Ray K, Potdar VA, Cherian SS, Pawar SD, Jadhav SM, Waregaonkar SR, et al. Characterization of the complete genome of influenza A (H5N1) virus isolated during the 2006 outbreak in poultry in India. Virus Genes. 2008;36:345–53. DOIPubMedGoogle Scholar
- Chakrabarti AK, Pawar SD, Cherian SS, Koratkar SS, Jadhav SM, Pal B, et al. Characterization of the influenza A H5N1 viruses of the 2008-09 outbreaks in India reveals a third introduction and possible endemicity. PLoS One. 2009;4:
e7846 . DOIPubMedGoogle Scholar - Nagarajan S, Tosh C, Smith DK, Peiris JSM, Murugkar HV, Sridevi R, et al. Avian influenza (H5N1) virus of clade 2.3.2 in domestic poultry in India. PLoS One. 2012;7:
e31844 . DOIPubMedGoogle Scholar - Tosh C, Nagarajan S, Kumar M, Murugkar HV, Venkatesh G, Shukla S, et al. Multiple introductions of a reassortant H5N1 avian influenza virus of clade 2.3.2.1c with PB2 gene of H9N2 subtype into Indian poultry. Infect Genet Evol. 2016;43:173–8. DOIPubMedGoogle Scholar
- Pawar SD, Kale SD, Rawankar AS, Koratkar SS, Raut CG, Pande SA, et al. Avian influenza surveillance reveals presence of low pathogenic avian influenza viruses in poultry during 2009-2011 in the West Bengal State, India. Virol J. 2012;9:151. DOIPubMedGoogle Scholar
- Potdar V, Hinge D, Satav A, Simões EAF, Yadav PD, Chadha MS. Laboratory-confirmed avian influenza A(H9N2) virus infection, India, 2019. Emerg Infect Dis. 2019;25:2328–30. DOIPubMedGoogle Scholar
- Kreman T. Capacity building for avian influenza H5N1. Centers for Disease Control and Prevention training presentation; October 6–10, 2007; Pune, India.
- World Health Organization. WHO information for molecular diagnosis of influenza virus—update. 2014. [cited on 2015 Nov 24] https://cdn.who.int/media/docs/default-source/influenza/global-influenza-surveillance-and-response-system/related-documents/molecular_diagnosis_influenza_virus_humans_update_201403.pdf
- Nguyen T, Rivailler P, Davis CT, Hoa T, Balish A, Dang NH, et al. Evolution of highly pathogenic avian influenza (H5N1) virus populations in Vietnam between 2007 and 2010. Virology. 2012;432:405–16. DOIPubMedGoogle Scholar
- Marinova-Petkova A, Franks J, Tenzin S, Dahal N, Dukpa K, Dorjee J, et al. Highly pathogenic reassortant avian influenza A(H5N1) virus clade 2.3.2.1a in poultry, Bhutan. Emerg Infect Dis. 2016;22:2137–41. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. H5N1 genetic changes inventory: a tool for international surveillance 2021 [cited on 2021 Jul 29]. https://www.cdc.gov/flu/avianflu/h5n1-genetic-changes.htm
- Long JS, Howard WA, Núñez A, Moncorgé O, Lycett S, Banks J, et al. The effect of the PB2 mutation 627K on highly pathogenic H5N1 avian influenza virus is dependent on the virus lineage. J Virol. 2013;87:9983–96. DOIPubMedGoogle Scholar
- Li J, Ishaq M, Prudence M, Xi X, Hu T, Liu Q, et al. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009;144:123–9. DOIPubMedGoogle Scholar
- Adlhoch C, Fusaro A, Gonzales JL, Kuiken T, Marangon S, Niqueux É, et al.; European Food Safety Authority; European Centre for Disease Prevention and Control and European Union Reference Laboratory for Avian Influenza. Avian influenza overview December 2020 - February 2021. EFSA J. 2021;19:
e06497 .PubMedGoogle Scholar - Verma N, Pooniya V, Kumar A. Clinical profile and outcome of influenza A/H1N1 in pediatric oncology patients during the 2015 outbreak: a single center experience from northern India. J Pediatr Hematol Oncol. 2017;39:e357–8. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: May 13, 2022
Table of Contents – Volume 28, Number 6—June 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Priya Abraham, ICMR-National Institute of Virology, Department of Health Research, Ministry of Health & Family Welfare, 20-A Dr. Ambedkar Road, Pune 411001, India
Top