Volume 28, Number 8—August 2022
Dispatch
Novel Chronic Anaplasmosis in Splenectomized Patient, Amazon Rainforest
Figure 1
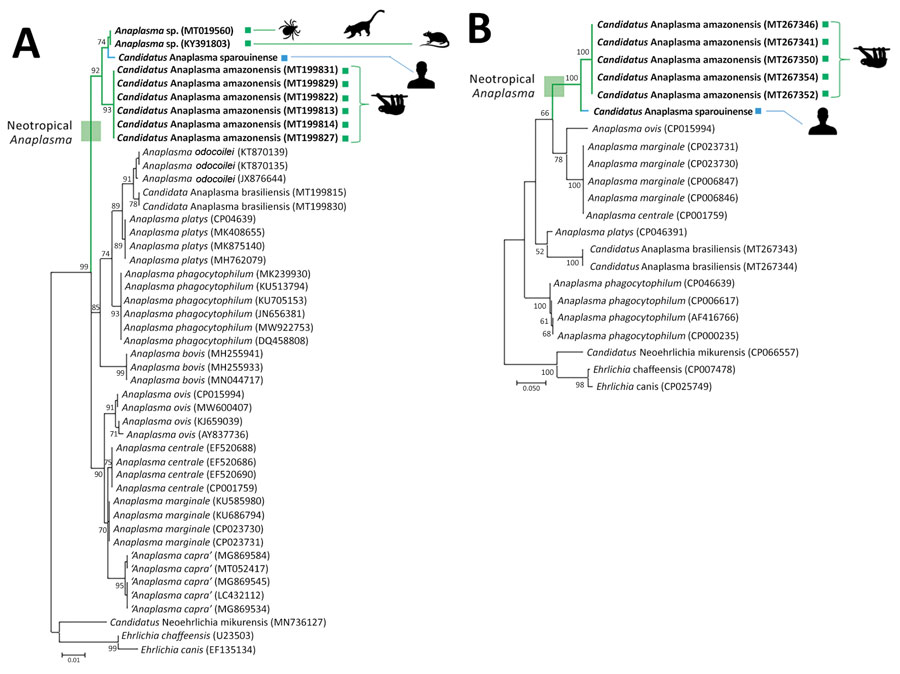
Figure 1. Anaplasma phylogenies for the Candidatus Anaplasma sparouinense species characterized from French Guiana and reference sequences. Trees were constructed by using maximum-likelihood estimations based on best-fit approximation for the evolutionary model Hasegawa-Kishino-Yano plus invariant sites for 16S rDNA with 485 unambiguously aligned bp (A) and ITS2 sequences with 387 unambiguously aligned bp (B). Bold indicates Anaplasma species and strains specific to the Neotropics. GenBank accession numbers of sequences used in analyses are shown on the phylogenetic trees. Numbers at nodes indicate percentage support of 1,000 bootstrap replicates. The scale bar is in units of substitution per site.
Page created: June 09, 2022
Page updated: July 20, 2022
Page reviewed: July 20, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.