Volume 29, Number 11—November 2023
Research
Genotypic Evolution of Klebsiella pneumoniae Sequence Type 512 during Ceftazidime/Avibactam, Meropenem/Vaborbactam, and Cefiderocol Treatment, Italy
Figure 3
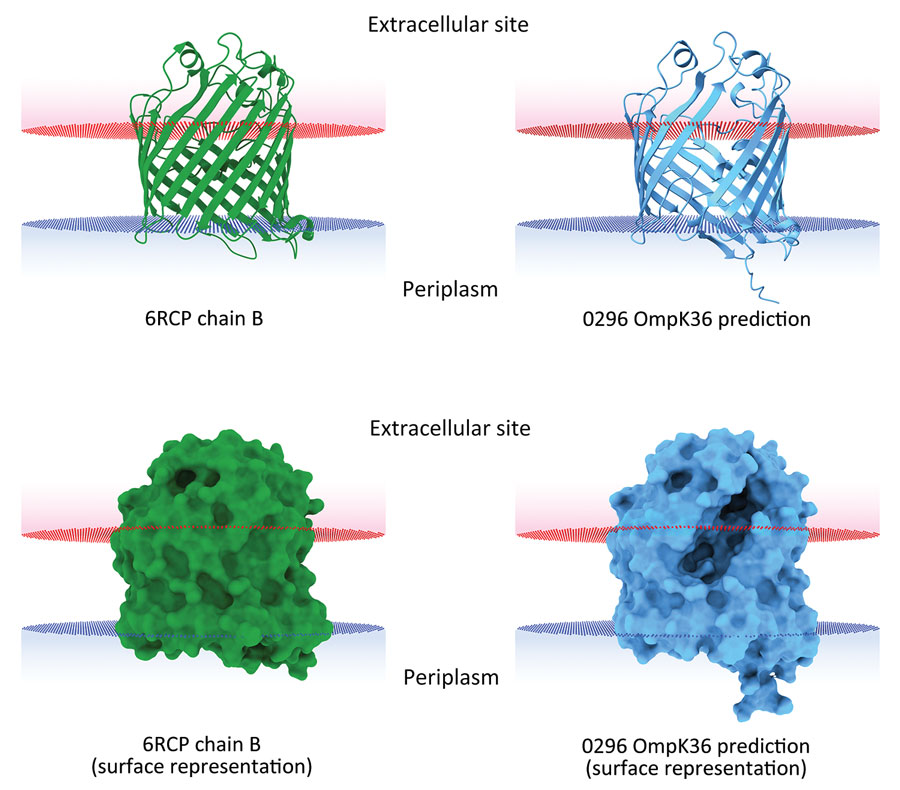
Figure 3. In silico 3-dimensional structure predictions for mutated OmpK36 porin in Klebsiella pneumoniae sequence type 512 strain 0296 in study of K. pneumoniae genotypic evolution during ceftazidime/avibactam, meropenem/vaborbactam, and cefiderocol treatment, Italy. The outer membrane porin OmpK36 from strain 0296 (blue) containing a 26 aa deletion from residue Thr263 through residue Tyr289 was modeled and compared with the model of reference OmpK36 chain B crystal structure from the Protein Data Bank (no. 6RCP; https://www.rcsb.org) (30). Both ribbon cartoon (top) and surface (bottom) models are shown. Structures for strain 0296 were obtained by using Alphafold2 on the European Galaxy server (https://usegalaxy.eu). Spatial arrangements of the porins in lipid bilayers were visualized by using the positioning of proteins in membranes web server in the Orientations of Proteins in Membranes database (31).
References
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al.; Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–55. DOIPubMedGoogle Scholar
- Shirley M. Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78:675–92. DOIPubMedGoogle Scholar
- Yahav D, Giske CG, Grāmatniece A, Abodakpi H, Tam VH, Leibovici L. New β-lactam-β-lactamase inhibitor combinations. Clin Microbiol Rev. 2020;34:e00115–20. DOIPubMedGoogle Scholar
- El-Lababidi RM, Rizk JG. Cefiderocol: a siderophore cephalosporin. Ann Pharmacother. 2020;54:1215–31. DOIPubMedGoogle Scholar
- Ito A, Nishikawa T, Matsumoto S, Yoshizawa H, Sato T, Nakamura R, et al. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60:7396–401. DOIPubMedGoogle Scholar
- David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al.; EuSCAPE Working Group; ESGEM Study Group. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–29. DOIPubMedGoogle Scholar
- Di Pilato V, Errico G, Monaco M, Giani T, Del Grosso M, Antonelli A, et al.; AR-ISS Laboratory Study Group on carbapenemase-producing Klebsiella pneumoniae. The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J Antimicrob Chemother. 2021;76:355–61. DOIPubMedGoogle Scholar
- Carattoli A, Arcari G, Bibbolino G, Sacco F, Tomolillo D, Di Lella FM, et al. Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2021;65:
e0057421 . DOIPubMedGoogle Scholar - Hobson CA, Pierrat G, Tenaillon O, Bonacorsi S, Bercot B, Jaouen E, et al. Klebsiella pneumoniae carbapenemase variants resistant to ceftazidime-avibactam: an evolutionary overview. Antimicrob Agents Chemother. 2022;66:
e0044722 . DOIPubMedGoogle Scholar - Karakonstantis S, Rousaki M, Kritsotakis EI. Cefiderocol: systematic review of mechanisms of resistance, heteroresistance and in vivo emergence of resistance. Antibiotics (Basel). 2022;11:723. DOIPubMedGoogle Scholar
- McElheny CL, Fowler EL, Iovleva A, Shields RK, Doi Y. In vitro evolution of cefiderocol resistance in an NDM-producing Klebsiella pneumoniae due to functional loss of CirA. Microbiol Spectr. 2021;9:
e0177921 . DOIPubMedGoogle Scholar - Fröhlich C, Sørum V, Tokuriki N, Johnsen PJ, Samuelsen Ø. Evolution of β-lactamase-mediated cefiderocol resistance. J Antimicrob Chemother. 2022;77:2429–36. DOIPubMedGoogle Scholar
- Lan P, Lu Y, Jiang Y, Wu X, Yu Y, Zhou J. Catecholate siderophore receptor CirA impacts cefiderocol susceptibility in Klebsiella pneumoniae. Int J Antimicrob Agents. 2022;60:
106646 . DOIPubMedGoogle Scholar - Klein S, Boutin S, Kocer K, Fiedler MO, Störzinger D, Weigand MA, et al. Rapid development of cefiderocol resistance in carbapenem-resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cirA. Clin Infect Dis. 2022;74:905–8. DOIPubMedGoogle Scholar
- Jousset AB, Poignon C, Yilmaz S, Bleibtreu A, Emeraud C, Girlich D, et al. Rapid selection of a cefiderocol-resistant Escherichia coli producing NDM-5 associated with a single amino acid substitution in the CirA siderophore receptor. J Antimicrob Chemother. 2023;78:1125–7. DOIPubMedGoogle Scholar
- Freire B, Ladra S, Parama JR. Memory-efficient assembly using Flye. IEEE/ACM Trans Comput Biol Bioinform. 2022;19:3564–77.
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–2. DOIPubMedGoogle Scholar
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42(D1):D206–14. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. DOIPubMedGoogle Scholar
- Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:
e000093 . DOIPubMedGoogle Scholar - Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. DOIPubMedGoogle Scholar
- Lam MMC, Wick RR, Judd LM, Holt KE, Wyres KL. Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microb Genom. 2022;8:
000800 . DOIPubMedGoogle Scholar - Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. DOIPubMedGoogle Scholar
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. DOIPubMedGoogle Scholar
- Wong JLC, Romano M, Kerry LE, Kwong HS, Low WW, Brett SJ, et al. OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat Commun. 2019;10:3957. DOIPubMedGoogle Scholar
- Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–5. DOIPubMedGoogle Scholar
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2014;111:4988–93. DOIPubMedGoogle Scholar
- García-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, et al. An ertapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother. 2010;54:4178–84. DOIPubMedGoogle Scholar
- Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, et al. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1997;41:699–701. DOIPubMedGoogle Scholar
- Cannatelli A, Giani T, D’Andrea MM, Di Pilato V, Arena F, Conte V, et al.; COLGRIT Study Group. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58:5696–703. DOIPubMedGoogle Scholar
- Verhamme DT, Arents JC, Postma PW, Crielaard W, Hellingwerf KJ. Glucose-6-phosphate-dependent phosphoryl flow through the Uhp two-component regulatory system. Microbiology (Reading). 2001;147:3345–52. DOIPubMedGoogle Scholar
- Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. 2018;4:
e000196 . DOIPubMedGoogle Scholar - Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother. 2017;61:e02097–16.PubMedGoogle Scholar
- Tooke CL, Hinchliffe P, Bonomo RA, Schofield CJ, Mulholland AJ, Spencer J. Natural variants modify Klebsiella pneumoniae carbapenemase (KPC) acyl-enzyme conformational dynamics to extend antibiotic resistance. J Biol Chem. 2021;296:
100126 . DOIPubMedGoogle Scholar - Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3:
e000110 . DOIPubMedGoogle Scholar - Zhang Z, Du W, Wang M, Li Y, Su S, Wu T, et al. Contribution of the colicin receptor CirA to biofilm formation, antibotic resistance, and pathogenicity of Salmonella Enteritidis. J Basic Microbiol. 2020;60:72–81. DOIPubMedGoogle Scholar
- Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24:944–56. DOIPubMedGoogle Scholar
- Zheng D, Bergen PJ, Landersdorfer CB, Hirsch EB. Differences in fosfomycin resistance mechanisms between Pseudomonas aeruginosa and Enterobacterales. Antimicrob Agents Chemother. 2022;66:
e0144621 . DOIPubMedGoogle Scholar - Ortiz-Padilla M, Portillo-Calderón I, de Gregorio-Iaria B, Blázquez J, Rodríguez-Baño J, Pascual A, et al. Interplay among different fosfomycin resistance mechanisms in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2021;65:e01911–20. DOIPubMedGoogle Scholar
- Thorpe HA, Booton R, Kallonen T, Gibbon MJ, Couto N, Passet V, et al. A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings. Nat Microbiol. 2022;7:2054–67. DOIPubMedGoogle Scholar
- Arcari G, Di Lella FM, Bibbolino G, Mengoni F, Beccaccioli M, Antonelli G, et al. A multispecies cluster of VIM-1 carbapenemase-producing Enterobacterales linked by a novel, highly conjugative, and broad-host-range IncA plasmid forebodes the reemergence of VIM-1. Antimicrob Agents Chemother. 2020;64:e02435–19. DOIPubMedGoogle Scholar
- Liu J, Wang R, Fang M. Clinical and drug resistance characteristics of Providencia stuartii infections in 76 patients. J Int Med Res. 2020;48:
300060520962296 . DOIPubMedGoogle Scholar - Molnár S, Flonta MMM, Almaş A, Buzea M, Licker M, Rus M, et al. Dissemination of NDM-1 carbapenemase-producer Providencia stuartii strains in Romanian hospitals: a multicentre study. J Hosp Infect. 2019;103:165–9. DOIPubMedGoogle Scholar
- Akbiyik A, Hepçivici Z, Eşer I, Uyar M, Çetin P. The effect of oropharyngeal aspiration before position change on reducing the incidence of ventilator- associated pneumonia. Eur J Clin Microbiol Infect Dis. 2021;40:615–22. DOIPubMedGoogle Scholar