Neoehrlichiosis in Symptomatic Immunocompetent Child, South Africa
Colleen Bamford
1
, Lucille H. Blumberg, Michelle Bosman, John Frean, Kim G.P. Hoek, Janet Miles, Charlotte Sriruttan, Ilse Vorster, and Marinda C. Oosthuizen
1
Author affiliations: Pathcare, East London, South Africa (C. Bamford); University of Cape Town, Cape Town, South Africa (C. Bamford); National Institute for Communicable Diseases, Johannesburg, South Africa (L.H. Blumberg, J. Frean, C. Sriruttan); University of Pretoria, Onderstepoort, South Africa (L.H. Blumberg, J. Frean, I. Vorster, M.C. Oosthuizen); University of Stellenbosch, Tygerberg, South Africa (L.H. Blumberg, K.G.P. Hoek); Ampath, East London, South Africa (M. Bosman); University of the Witwatersrand, Johannesburg, South Africa (J. Frean); PathCare Reference Laboratory, Cape Town (K.G.P. Hoek); East London, South Africa (J. Miles)
Main Article
Figure
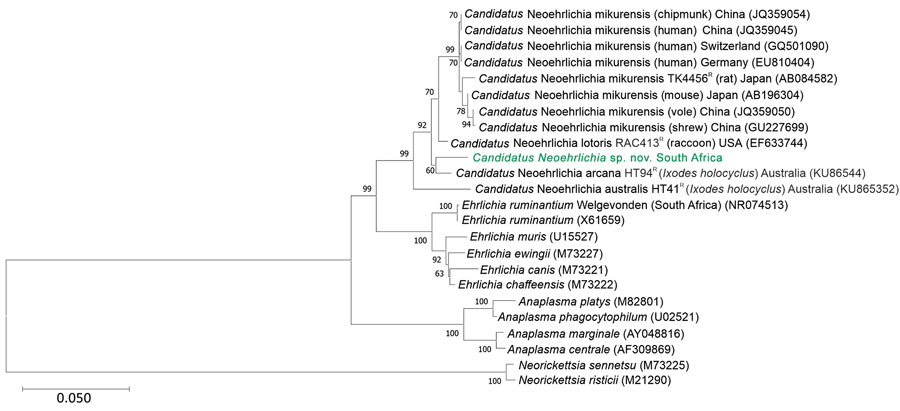
Figure. Phylogenetic tree of Candidatus Neoehrlichia species detected in a case of neoehrlichiosis in a symptomatic immunocompetent child, South Africa. We used Kimura 2-parameter plus gamma plus invariable site substitution model in MEGA X (https://www.megasoftware.net) to infer a maximum-likelihood phylogenetic tree in combination with the bootstrap method using 1,000 replicates. Green bold text indicates Candidatus Neoehrlichia species detected from the patient in this case-study; we compared this isolate to other Anaplasmataceae species detected from ticks, humans, and mammals available in GenBank (indicated by species name and GenBank accession number; host species and country are given for other Candidatus Neoehrlichia species). Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: December 22, 2022
Page updated: January 21, 2023
Page reviewed: January 21, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
