Volume 29, Number 4—April 2023
Research Letter
Powassan Virus Infection Detected by Metagenomic Next-Generation Sequencing, Ohio, USA
Figure
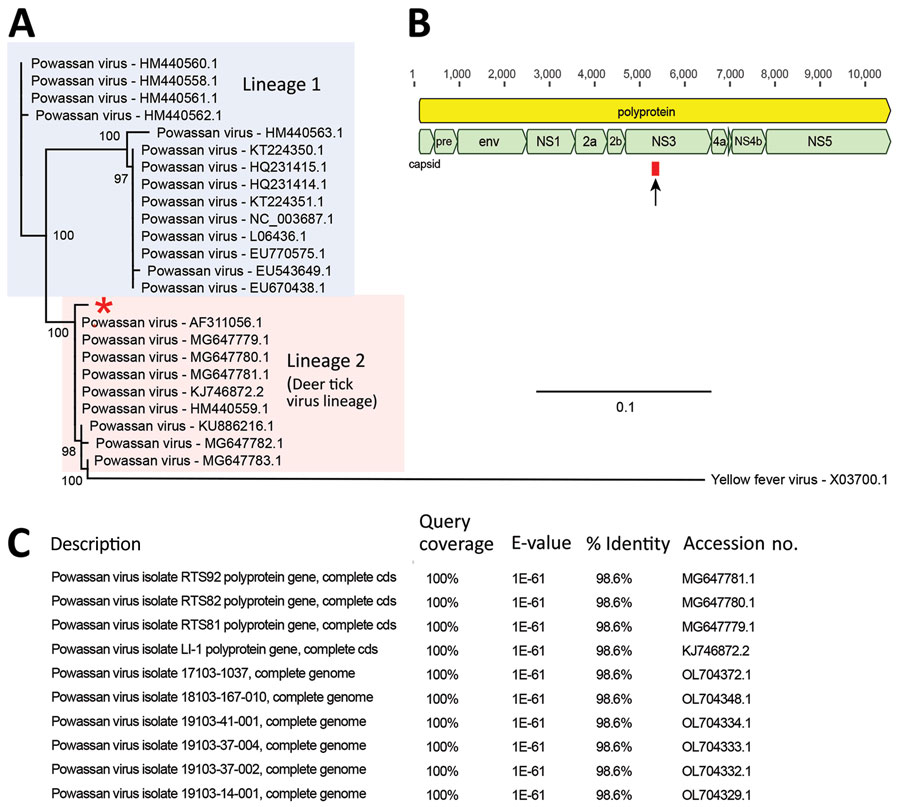
Figure. Identification and phylogenetic analysis of Powassan virus found in cerebrospinal fluid of 4-year-old boy detected by metagenomic next-generation sequencing, Ohio, USA. A) Phylogenetic analysis of the 140-bp region in the Powassan virus genome corresponding to the single sequence read detected by metagenomic next-generation sequencing. Single read from the patient in this study was aligned with sequences from 23 representative Powassan virus genomes from lineage 1 (blue shaded box) and lineage 2 (deer tick virus lineage, pink shaded box) and 1 yellow fever virus sequence as an outgroup by using MAFFT v7.388 (Appendix reference 11 ). Phylogenetic tree was constructed by using the maximum-likelihood method and PhyML 3.0 software (Appendix reference 12); support values for the main branches are shown. Powassan virus from our patient (red asterisk) belongs to lineage 2. GenBank accession numbers are shown for each sequence. Scale bar indicates nucleotide substitutions per site. B) Powassan virus genome showing major capsid and nonstructural genes. Single sequence read from the patient mapped to the NS3 gene (arrow and red box). C) List of top 10 GenBank reference sequences matching the patient’s 140-nt read after using MegaBLAST (https://blast.ncbi.nlm.nih.gov) alignment as default setting, each showing 98.6% sequence identity. If Powassan virus sequences were excluded from the reference database, no other matches in GenBank were found. Cds, coding sequence; env, envelope protein; NS, nonstructural; pre, M protein precursor peptide.
1Current affiliation: St. Louis Children’s Hospital, St. Louis, Missouri, USA.