Volume 29, Number 5—May 2023
Research Letter
New Genotype of Coxiella burnetii Causing Epizootic Q Fever Outbreak in Rodents, Northern Senegal
Figure
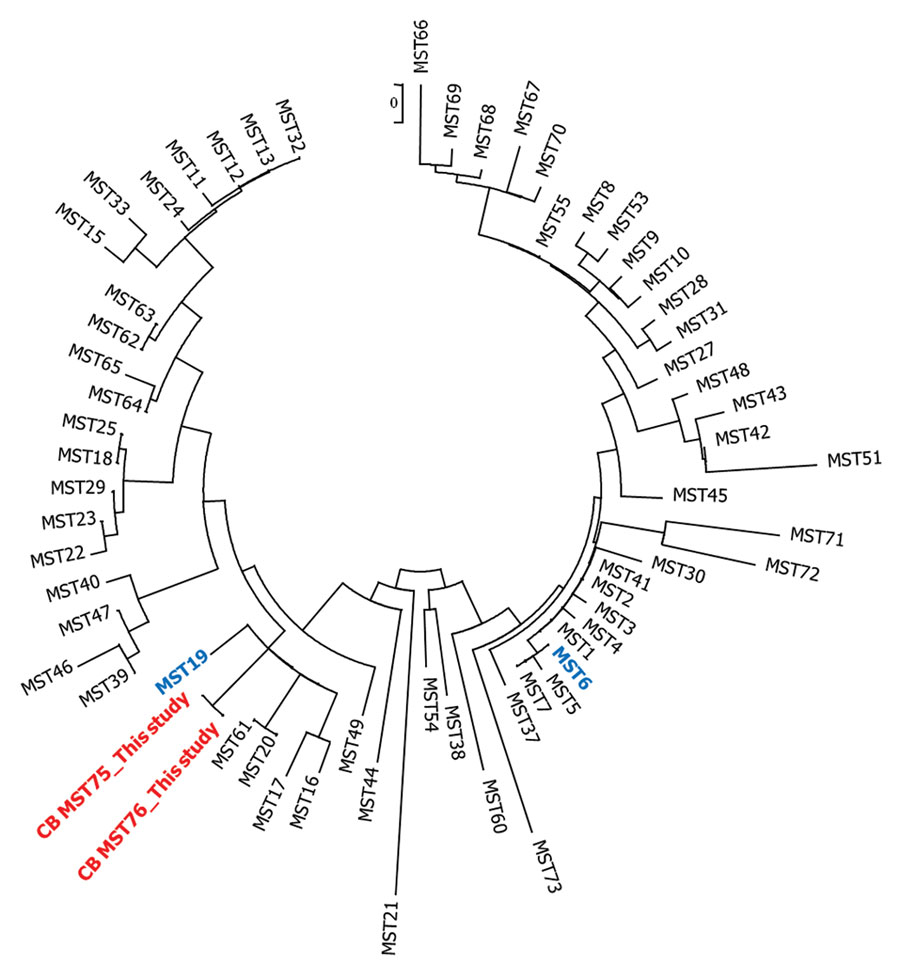
Figure. Neighbor-joining circular unrooted tree showing the relationship between Coxiella burnetii genotypes described in study of new genotype of C. burnetii causing epizootic Q fever outbreak in rodents, northern Senegal. MST75 and MST76 (red) were compared with genotypes already found in Senegal, MST19 and MST6 (blue), and other genotypes. The analysis involved 64 nt sequences. All positions containing gaps and missing data were eliminated. There were a total of 4,692 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (https://www.megasoftware.net). MST, multispacer sequence typing.
Page created: February 23, 2023
Page updated: April 19, 2023
Page reviewed: April 19, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.