Volume 29, Number 7—July 2023
Research Letter
Detection of Mycobacterium angelicum in Human Urinary Tract, French Polynesia
Abstract
We definitively characterized Mycobacterium angelicum, an aquatic zoonotic opportunistic pathogen of the M. szulgai complex, using a polyphasic approach that included whole-genome sequencing. The sequence was obtained on the island of Tahiti, French Polynesia, from a urine specimen collected from a patient experiencing a urinary tract infection.
Mycobacterium angelicum, a slow-growing mycobacterium associated with animals living in freshwater environments, was delineated within the M. szulgai complex by 16S rRNA gene sequencing after its initial isolation from a freshwater angelfish (Pterophyllum scalare) in 2003 (1). In line with international recommendations (2), M. angelicum was formally described in 2015 as a new species including the seminal 2003 isolate, 2 additional isolates from freshwater fish, and a fourth isolate recovered from a freshwater tank containing tortoises (1). Meanwhile, another isolate was identified in Benin from the rodent Crocidura olivieri (3). A clinical isolate recovered from a respiratory tract sample taken from a patient in Northern Ireland was also tentatively identified as M. angelicum on the basis of 16S rRNA gene sequencing (4). We report another clinical isolate identified as M. angelicum on the basis of a polyphasic identification approach including whole-genome sequencing (WGS).
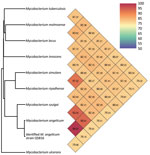
A middle-aged patient sought care for active struvite urolithiasis in the left kidney at the main hospital in Papeete, French Polynesia, 18 years after a right nephrectomy for obstructive pyonephrosis. We were unable to follow up with the patient beyond this medical episode. Of 3 successive urine samples collected over 3 consecutive days, which all lacked acid-fast bacilli after Ziehl-Neelsen staining, we successfully cultured 1 urine sample on MGIT (Becton, Dickinson and Company, https://www.bd.com) in 11 days. Culture on Löwenstein Jensen medium (Becton Dickinson) at 37°C under aerobic conditions remained negative after 3 months’ incubation. This isolate positively stained by Ziehl-Neelsen staining; it exhibited rod-shaped, pink-stained bacteria measuring 3.225 + 0.858 μm by 0.717 + 0.048 μm under electron microscopy observation using a SU5000 SEM electron microscope (Hitachi, https://www.hitachi.com). Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using a Microflex spectrometer and software (Bruker Daltonics, https://www.bruker.com), as previously described (5), yielded a noninformative score of 1.31; a derived dendrogram clustered the isolate within the M. szulgai group. We conducted WGS concatenating Illumina (https://www.illumina.com) and Nanopore (https://nanoporetech.com) reads using Spades software version 3.15.4, as previously described (6); this process yielded 66.4% guanine-cytosine content, 0.23% gap ratio, and a 6,673,592-bp sequence distributed into 51 contigs encoding for 5,707 proteins, 56 tRNA, 3 rRNA, and 2 CRISPRs with a 90.5% total coding ratio. We deposited the sequence into GenBank (submission identification no. 2639860). As a first step, BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) of the 1,326,459-bp longest contig yielded 98% coverage and 99.3% similarity with an environmental M. angelicum isolate strain DSM 45057 WGS (GenBank accession no. NZ_MVHE01000100) (1) and a 98.6% DNA–DNA hybridization using the Type Strain Genome Server (Leibniz Institute DSMZ, https://www.dsmz.de). As a second step, we used the Orthologous Average Nucleotide Identity tool version 0.93.1 (7); the isolate clustered with the best BLAST hit M. angelicum isolate with 99.73% genome similarity, whereas further genome sequence similarity values were 93.25% with M. szulgai, 93.01% with M. riyadhense, and >85% with other mycobacteria (Figure). Those data identified our isolate as the M. angelicum Tahiti strain; we deposited it into the Collection de Souches de l’Unité des Rickettsies (CSUR Q5816). No antimicrobial resistance–encoding sequences were predicted in the M. angelicum Tahiti strain genome. In agreement with the qualification of M. angelicum as a human pathogen by the Center for Genomic Epidemiology online platform (http://www.genomicepidemiology.org), 24 pathogenicity-associated genes were identified in the M. angelicum Tahiti strain, all highly conserved in Mycobacterium species (Appendix). The location of the ClpS gene in 74,165–74,377, its 97.14% sequence similarity with the homologous gene in Mycobacterium, and encoding an ATP-dependent Clp protease were predicted as nonpathogenicity factors affecting antimicrobial metabolism and rifampin resistance to protect the Mycobacterium cell wall against various stresses (8,9).
We identified the Tahiti strain as M. angelicum by combining WGS with phenotypic data in the presence of controls. The identification pathway was an opportunity to make a clinical M. angelicum WGS available; 1 M. angelicum partial genome sequence (GenBank NZ_MVHE01000100.1) derived from a freshwater angelfish isolate had been described previously (1). We did not find reports of M. angelicum as a contaminant in that study, in a urine collection device, or as a laboratory contaminant. Furthermore, we did not find previous reports of M. angelicum analysis in either of the 2 laboratories that handled this patient’s urine or the strain itself; we concluded that this M. angelicum isolate was not a contaminant. This isolate was the only microorganism we were able to isolate by culture from a urine sample; however, its role in the complex urinary tract pathology of this patient remained putative.
Previously, an isolate of M. angelicum was identified from a bronchoalveolar sample collected from a patient with chronic obstructive pulmonary disease in Northern Ireland; identification was based on 100% partial identity (817-bp) 16S rRNA gene sequencing with the reference (3). Our study underscores the need for WGS sequencing of bacterial pathogens not identified by first-line phenotypic schemes, including appropriate matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (5).
Mr. Keita is a PhD student in the Institut Hospitalo-Universitaire Méditerranée Infection, Marseille, France. His primary research interest is in the clinical microbiology of mycobacteria of tropical sources, including Mycobacterium ulcerans. Dr. Morsli is an expert in the genomics and metagenomics of mycobacteria, working at the Centre Hospitalier Universitaire, Nîmes, France.
Acknowledgments
According to French law, anonymous case report does not require specific ethical approval when the patient does not oppose reporting.
M.L.K. is supported by the Fondation Meditérranée Infection. This work was supported by Microbes, Evolution, Phylogeny and Infections, Aix Marseille University, Institut de Recherche pour le Développement.
Authors’ contributions: M.L.K. and M.M.: data collection, data cleaning, design of the study, data interpreting, validation and writing of the manuscript. M.L., G.B., and C.V.: clinical examination, data collection, validation, interpretation, and writing of the manuscript. M.D.: design of the study, data interpretation, validation, funding, critical review of the manuscript, coordination and direction of the work. All authors declare that they have read and approved the manuscript.
References
- Pourahmad F, Pate M, Ocepek M, Borroni E, Cabibbe AM, Capitolo E, et al. Mycobacterium angelicum sp. nov., a non-chromogenic, slow-growing species isolated from fish and related to Mycobacterium szulgai. Int J Syst Evol Microbiol. 2015;65:4724–9. DOIPubMedGoogle Scholar
- Christensen H, Bisgaard M, Frederiksen W, Mutters R, Kuhnert P, Olsen JE. Is characterization of a single isolate sufficient for valid publication of a new genus or species? Proposal to modify recommendation 30b of the Bacteriological Code (1990 Revision). Int J Syst Evol Microbiol. 2001;51:2221–5. DOIPubMedGoogle Scholar
- Durnez L, Suykerbuyk P, Nicolas V, Barrière P, Verheyen E, Johnson CR, et al. Terrestrial small mammals as reservoirs of Mycobacterium ulcerans in benin. Appl Environ Microbiol. 2010;76:4574–7. DOIPubMedGoogle Scholar
- Davies E, Wieboldt J, Stanley T, Maeda Y, Smyth M, Stanley S, et al. Isolation and identification of ‘Mycobacterium angelicum’ from a patient with type II respiratory failure: suggested reporting guidelines to molecular clinical laboratories. Br J Biomed Sci. 2012;69:134–6. DOIPubMedGoogle Scholar
- Robinne S, Saad J, Morsli M, Hamidou ZH, Tazerart F, Drancourt M, et al. Rapid identification of Mycobacterium tuberculosis complex using mass spectrometry: a proof of concept. Front Microbiol. 2022;13:
753969 . DOIPubMedGoogle Scholar - Morsli M, Boudet A, Kerharo Q, Stephan R, Salipante F, Dunyach-Remy C, et al. Real-time metagenomics-based diagnosis of community-acquired meningitis: A prospective series, southern France. EBioMedicine. 2022;84:
104247 . DOIPubMedGoogle Scholar - Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–3. DOIPubMedGoogle Scholar
- Adilijiang G, Feng S, Mi K, Deng H. [Quantitative proteomics analysis of ClpS-mediated rifampicin resistance in Mycobacterium.] [in Chinese]. Sheng Wu Gong Cheng Xue Bao. 2014;30:1115–27.PubMedGoogle Scholar
- Marsee JD, Ridings A, Yu T, Miller JM. Mycobacterium tuberculosis ClpC1 N-terminal domain is dispensable for adaptor protein-dependent allosteric regulation. Int J Mol Sci. 2018;19:3651. DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: June 12, 2023
1These authors contributed equally to this article.
Table of Contents – Volume 29, Number 7—July 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Michel Drancourt, MEPHI, IHU Méditerranée Infection, 19-21, Blvd Jean Moulin, 13005 Marseille CEDEX 05, France
Top