Six Extensively Drug-Resistant Bacteria in an Injured Soldier, Ukraine
Patrick T. Mc Gann, Francois Lebreton, Brendan T. Jones, Henry D. Dao, Melissa J. Martin, Messiah J. Nelson, Ting Luo, Andrew C. Wyatt, Jason R. Smedberg, Joanna M. Kettlewell, Brain M. Cohee, Joshua S. Hawley-Molloy, and Jason W. Bennett
Author affiliations: Multidrug-Resistant Organism Repository and Surveillance Network, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA (P.T. Mc Gann, F. LeBreton, B.T. Jones, H.D. Dao, M.J. Martin, M.J. Nelson, T. Luo, J.W. Bennett); Landstuhl Regional Medical Center, Landstuhl, Germany (A.C. Wyatt, J.R. Smedberg, J.M. Kettlewell, J.S. Hawley-Malloy); 512th Field Hospital, Rhine Ordinance Barracks, Germany (B.M. Cohee)
Main Article
Figure
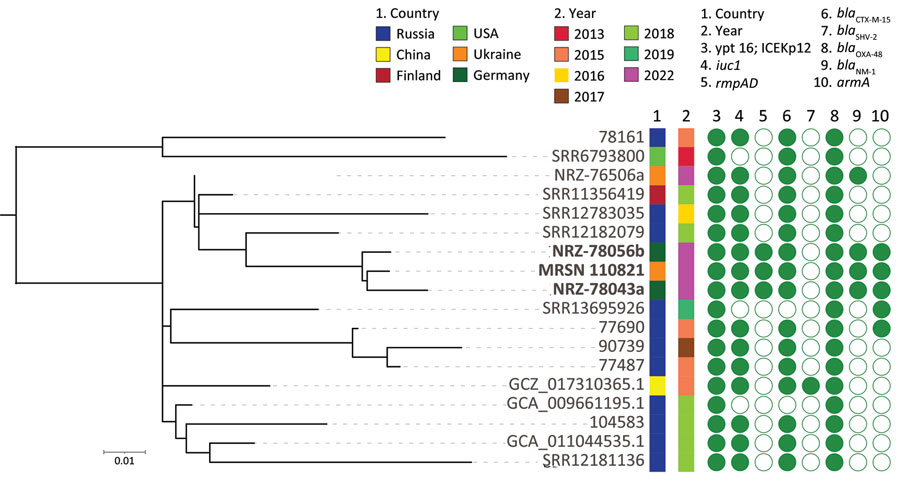
Figure. Core genome, SNP-based phylogenetic tree for Klebsiella pneumoniae from an injured service member from Ukraine (MRSN 110821) and 17 closely related sequence type 395 K. pneumoniae. In addition to MRSN 110821, the dataset included 14 subclade B2 isolates and 3 NDM-1/OXA-48–producing isolates available in public databases. Country of origin, year of collection, and presence (closed circle) or absence (open circle) of selected virulence and antimicrobial resistance genes are indicated. The midpoint was used as a root for the phylogenetic tree. K. pneumoniae MRSN 110821 from this study and the 2 highly related strains from Germany are highlighted in boldface. Scale bar indicates the ratio of substitutions per site for a 1,665 bp alignment of variable sites in the core genomes of the 18 strains.
Main Article
Page created: June 29, 2023
Page updated: July 20, 2023
Page reviewed: July 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
