Imported Cholera Cases, South Africa, 2023
Anthony M. Smith

, Phuti Sekwadi, Linda K. Erasmus, Christine C. Lee, Steven G. Stroika, Sinenhlanhla Ndzabandzaba, Vinitha Alex, Jeremy Nel, Elisabeth Njamkepo, Juno Thomas, and François-Xavier Weill
Author affiliations: University of Pretoria, Pretoria, South Africa (A.M. Smith); National Institute for Communicable Diseases, Johannesburg, South Africa (A.M. Smith, P. Sekwadi, L.K. Erasmus, J. Thomas); US Centers for Disease Control and Prevention, Atlanta, Georgia, USA (C.C. Lee, S.G. Stroika); National Heath Laboratory Service, Johannesburg (S. Ndzabandzaba, V. Alex); University of the Witwatersrand, Johannesburg (S. Ndzabandzaba, V. Alex, J. Nel); Institut Pasteur, Université Paris Cité, Paris, France (E. Njamkepo, F.-X. Weill)
Main Article
Figure
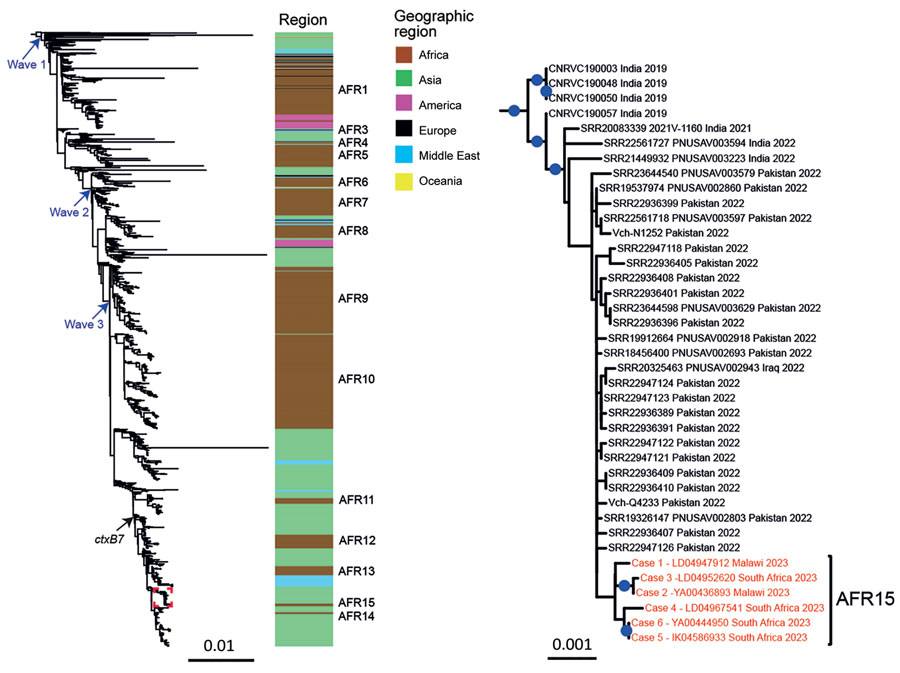
Figure. Maximum-likelihood phylogeny of Vibrio cholerae O1 El Tor isolates collected in South Africa, 2023, compared with 1,443 reference seventh pandemic V. cholerae El Tor (7PET) genomic sequences. A6 was used as the outgroup. The genomic waves and acquisition of the ctxB7 allele are indicated. Color coding indicates the geographic origins of the isolates; sublineages previously introduced into Africa (AFR1, AFR3–AFR14) are shown at right. A magnification of the clade containing the 6 isolates from South Africa (red text) is shown at right. For each genome, name (or accession number), country where contamination occurred, and year of sample collection are shown at the tips of the tree. The 6 isolates collected in South Africa belong to a new 7PET wave 3 sublineage called AFR15. Blue dots indicate bootstrap values ≥90%. Scale bars indicate number of nucleotide substitutions per variable site.
Main Article
Page created: June 15, 2023
Page updated: July 20, 2023
Page reviewed: July 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
