Volume 29, Number 9—September 2023
Dispatch
Reoccurring Escherichia coli O157:H7 Strain Linked to Leafy Greens–Associated Outbreaks, 2016–2019
Figure 1
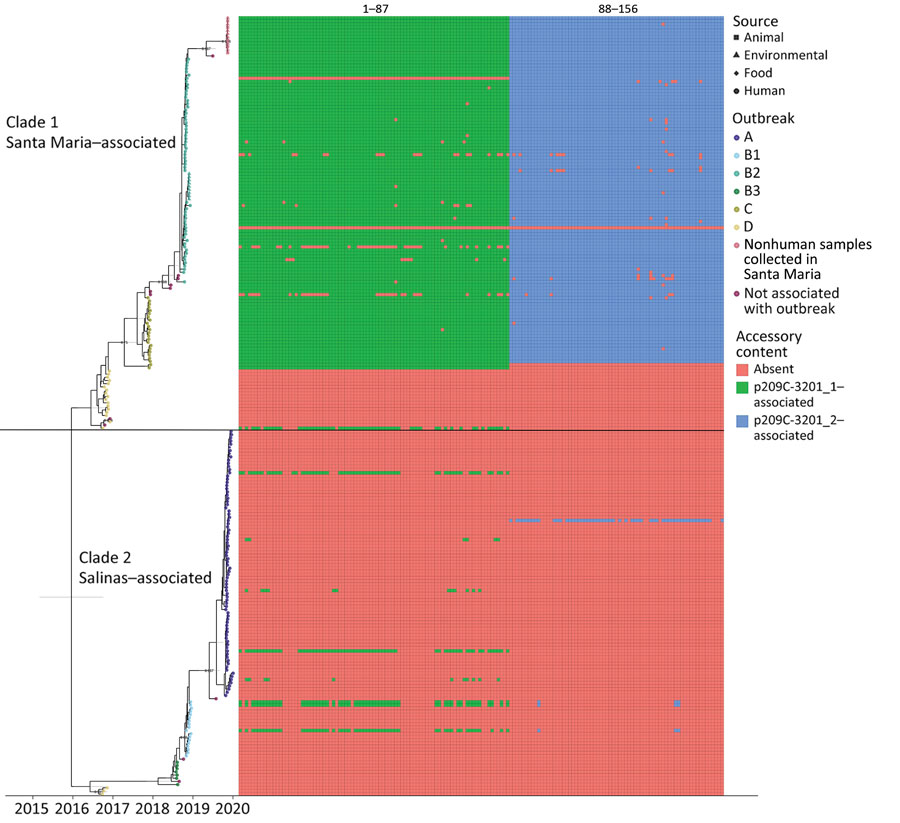
Figure 1. Tip-dated maximum clade credibility tree of 245 isolates of reoccurring Escherichia coli O157:H7 strain REPEXH02 linked to leafy greens–associated outbreaks, 2016–2019, generated in BEAST2 (https://www.beast2.org). Tips are aligned with the date of collection; calendar year is shown on the x-axis. Tips are colored according to the outbreak to which each isolate belonged; the shape corresponds to sample type (e.g., human, animal, environmental, or food). A horizontal black line segregates the two identified clades. Clade 1 contains outbreak B2 where some illness was traced back to Santa Maria, California, USA, as well as environmental samples collected in that region. Clade 2 contains outbreak A, which was traced back to the Salinas Valley, California. The presence/absence matrix to the right of the tree displays accessory genome content identified using Roary/scoary with 90% sensitivity and specificity to a subset of clade 1 isolates. A legend for accessory genome feature labels is included in Appendix 1 Table 5.
1These first authors contributed equally to this article.
2Current affiliation: Tulane National Primate Research Center, Covington, Louisiana, USA.