Volume 29, Number 9—September 2023
Dispatch
Fatal Necrotizing Enterocolitis in Neonate Caused by Cronobacter sakazakii Sequence Type 64 Strain of CRISPR Sublineage b
Abstract
We report fatal neonatal necrotizing enterocolitis in China caused by Cronobacter sakazakii capsular profile K1:CA1, sequence type 64, and CRISPR type 197. Phylodynamic analyses indicated that the strain originated from the ancient, widespread, and antimicrobial drug–sensitive CRISPR sublineage b. Enhanced surveillance and pathogenesis research on this organism are required.
Cronobacter sakazakii is a major foodborne pathogen that is associated with outbreaks of life-threatening necrotizing enterocolitis, meningitis, and sepsis in neonates and infants. Although the incidence of this pathogen is low, the case-fatality rate is high in premature and immunocompromised infants (1,2). Multilocus sequence typing (MLST) is a powerful tool for effectively identifying and discriminating different Cronobacter strains. Specific sequence types (STs) and clonal complexes are closely related to infections (3).
Compared with MLST, CRISPR (clustered regularly interspaced short palindromic repeats) typing is superior for distinguishing similar strains (4). C. sakazakii ST64, the major ST in food samples, was further divided into 2 sublineages based on CRISPR diversity (5). We report a C. sakazakii ST64 strain that caused necrotizing enterocolitis in a neonate in China and further examine its origin and phylogenetic relationship with ST64 strains based on CRISPR diversity and whole-genome single-nucleotide polymorphism (wgSNP).
This study was approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center (Guangzhou, China; no. 2016081029). Experiments were performed at the Institute of Microbiology, Guangdong Academy of Sciences, and analyses and manuscript preparation were completed at Guangdong University of Technology.
On April 28, 2019, a 17-day-old male neonate born with severe congenital heart disease and perioral cyanosis for 2 hours was hospitalized in a children’s hospital in Guangzhou, China. The patient had necrotizing enterocolitis symptoms develop on May 6 and was given meropenem and metronidazole as antiinfection therapy. However, his symptoms did not improve, and intestinal perforation and peripheral hydrocephalus developed a few days later. Despite the efforts of the doctor, the patient died.
We identified a Cronobacter species isolated from ascites by using an automated VITEK 2 Compact system (bioMérieux, https://www.biomerieux.com). An isolate, GZfs, was identified as C. sakazakii ST64 of serotype O2. This ST has not previously been reported to cause neonatal necrotizing enterocolitis (6). C. sakazakii GZfs were susceptible to almost all antimicrobial drugs tested, except cephalothin (Appendix Table). We sequenced genomic and plasmid DNA by using the PacBio RS II (Pacific Biosciences, https://www.pacb.com) and HiSeq (Illumina, https://www.illumina.com) platforms, assembled, and annotated as described (1,7). C. sakazakii GZfs had a single circular chromosome, 4.2 Mb, 57.11% GC content, and 2 plasmids (denoted as pFS1, 115,925 bp, 57.09% GC; and pFS2, 110,391 bp, 50.10% GC) (GenBank accession nos. CP123201‒3).
In our previous study, we divided C. sakazakii ST64 strains into 2 CRISPR sublineages, a and b, and compared antimicrobial drug resistance profile strains in sublineage a with strains in sublineage b (5). To explore the origin of this pathogenic strain and its phylogenetic relationship with other C. sakazakii ST64 strains, we performed whole-genome sequencing of 9 ST64 strains (GenBank accession nos. JARUQD000000000‒ L000000000) and downloaded all ST64 strains with whole-genome sequences from the Cronobacter PubMLST database (https://pubmlst.org/organisms/cronobacter-spp) and GenBank genome databases. We provide antimicrobial drug resistance results of 14 food-source ST64 strains (Appendix Table).
After deleting all poor-quality sequences and duplicate strains, we used 66 whole-genome sequences for further analyses. We extracted CRISPR arrays and spacers from those sequences and assigned CRISPR type (CT) numbers to 55 strains with intact CRISPR arrays, according to methods from our previous study (4). All ST64 strains had the same 2 spacers: CRISPR3, which was not detected in our previous study because of the lack of cas genes (7), and CRISPR3, which was not useful for CT in this ST. There were 25 CTs, including 17 new, and we identified C. sakazakii GZfs as a new type of CT197 (Appendix). Based on spacer composition, GZfs belonged to CRISPR sublineage b. However, no other strain was found to have an identical spacer profile.
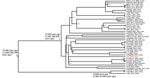
We calculated the wgSNP of ST64 strains by using Harvest software (8) and extracted those strains by using SNP-sites software (9). We constructed a maximum-likelihood phylogenetic tree by using FastTree software (10) and edited in iTOL (11). We used a Bayesian phylogenetic approach to estimate the nucleotide substitution rates and divergence times of C. sakazakii ST64 according to a previous study (7). The maximum-likelihood tree based on the wgSNPs of ST64 strains also showed 2 distinct phylogenetic clusters in accordance with the CRISPR sublineages (Figure 1). The strains in sublineage a were all from food sources.
Sublineage b contained more strains and diverse sources. Moreover, C. sakazakii GZfs and all clinical source strains (C. sakazakii KMB-550, MOD1-1121-73, and CDC1121-73) in public databases belonged to this cluster. C. sakazakii MOD1-1121-73 and CDC1121-73 were isolated from bronchial washes; there was no other patient or disease information regarding those clinical strains.
The genome-wide substitution rate of C. sakazakii ST64 was estimated to be 2.3 × 106 substitutions/site/year (95% CI 1.0 × 107–5.3 × 106 substitutions/site/year). According to the maximum clade credibility (MCC) tree (Figure 2), the likely most recent common ancestor of CRISPR sublineage b was 47,500 (95% CI 11,600–300,700) years ago, earlier than for sublineage a, which was 10,900 (95% CI 1,300–11,600) years ago. C. sakazakii GZfs had a relatively close phylogenetic relationship with the food-source C. sakazakii strain ZV-3645-16 in Slovenia; environmental strains C. sakazakii Crono01, Crono02-YL, and Crono03-YL; and the food-source strain C. sakazakii cro3825W in China (Figure 1, Figure 2). This finding indicates a close environment food-clinic relationship in dissemination.
We identified acquired drug resistance genes by using ResFinder 2.1. (https://cge.cbs.dtu.dk/services/ResFinder-2.1). All 3 strains in sublineage a acquired the antimicrobial resistance (AMR) genes tet(A)/aadA2/dfrA12/qacE/sul1, in accordance with their resistance to tetracycline and trimethoprim/sulfamethoxazole (Figure 1; Appendix Table). Five strains in sublineage b had AMR genes; 3 strains (C. sakazakii cro645A3-1, cro3040W, cro4114A1) and 1 strain (C. sakazakii cro4114B2) isolated from vegetables harbored the AMR genes blaTEM-116 and qnrA3. Four strains were susceptible to all tested antimicrobial drugs (Appendix Table). One environmental strain, C. sakazakii FDA1024695-210-001, had tet(B)/aph(3′′)-Ib/aph(6)-Id genes. A total of 92.1% (58/63) strains in sublineage b lacked AMR genes. All 4 clinical strains, including C. sakazakii GZfs, did not have AMR genes. In a previous study, C. sakazakii ST494 strain was sensitive to all antimicrobial drugs used for treatment; however, the patient died (12). Those results suggested that AMR might not be the major reason for the high case-fatality rate associated with this pathogenic infection. Both C. sakazakii ST494 and ST64 did not belong to the common pathogenic clonal complex. Enhanced surveillance and pathogenesis research of this organism are warranted.
The virulence genes in C. sakazakii remain unclear (13), and 2 T6SS and 1 prophage on the chromosome of GZfs might contribute to pathogenicity. The capsular profile of the GZfs was determined to be K1:CA1, as in our previous study (1), and plasmid pFS1 was closely related to the IncFIB-type virulence plasmid pESA3 identified in pathogenic C. sakazakii strains and pGW2 in C. sakazakii GZcsf-1, which causes meningitis in neonates (1,14). More attention should be given to the study of virulence and pathogenesis.
We report 1 C. sakazakii ST64 strain, GZfs, causing fatal neonatal necrotizing enterocolitis in China that did not belong to the previously identified common pathogenic clonal complexes or STs (3). It belongs to the ancient, widespread, and antimicrobial drug‒sensitive CRISPR cluster b of ST64. AMR might not be the major reason for the high case-fatality rate for this pathogen. Public health would benefit from identification of virulence genes and pathogenic mechanisms of C. sakazakii.
Dr. Zeng is an associate professor at Guangdong University of Technology, Guangzhou, China. Her primary research interests are adaptive evolution, drug resistance, and phage treatment of major pathogenic bacteria.
Acknowledgments
We thank Editage (https://www.editage.cn) for English language editing.
This study was supported by grants from the National Natural Science Foundation of China (32072327) and Guangdong Basic and Applied Basic Research Foundation (2021A1515010865).
References
- Zeng H, Lei T, He W, Zhang J, Liang B, Li C, et al. Novel multidrug-resistant Cronobacter sakazakii causing meningitis in neonate, China, 2015. Emerg Infect Dis. 2018;24:2121–4. DOIPubMedGoogle Scholar
- Taylor MG, Amerson-Brown MH, Hulten K, Cameron LH, Holzmann-Pazgal G, Edwards MS, et al. Two cases of Cronobacter sakazakii meningitis in infants: the importance of early advanced brain imaging and public health reporting. Pediatr Infect Dis J. 2021;40:e346–8. DOIPubMedGoogle Scholar
- Ogrodzki P, Forsythe SJ. DNA-sequence based typing of the Cronobacter genus Using MLST, CRISPR-cas array and capsular profiling. Front Microbiol. 2017;8:1875. DOIPubMedGoogle Scholar
- Zeng H, Li C, He W, Zhang J, Chen M, Lei T, et al. Cronobacter sakazakii, Cronobacter malonaticus, and Cronobacter dublinensis genotyping based on CRISPR locus diversity. Front Microbiol. 2019;10:1989. DOIPubMedGoogle Scholar
- Zeng H, Li C, Ling N, Zhang J, Chen M, Lei T, et al. Prevalence, genetic analysis and CRISPR typing of Cronobacter spp. isolated from meat and meat products in China. Int J Food Microbiol. 2020;321:
108549 . DOIPubMedGoogle Scholar - Gopinath GR, Chase HR, Gangiredla J, Eshwar A, Jang H, Patel I, et al. Genomic characterization of malonate positive Cronobacter sakazakii serotype O:2, sequence type 64 strains, isolated from clinical, food, and environment samples. Gut Pathog. 2018;10:11. DOIPubMedGoogle Scholar
- Zeng H, Zhang J, Wu Q, He W, Wu H, Ye Y, et al. Reconstituting the history of Cronobacter evolution driven by differentiated CRISPR activity. Appl Environ Microbiol. 2018;84:e00267–18. DOIPubMedGoogle Scholar
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. DOIPubMedGoogle Scholar
- Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:
e000056 . DOIPubMedGoogle Scholar - Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:
e9490 . DOIPubMedGoogle Scholar - Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. DOIPubMedGoogle Scholar
- Chaves CEV, Brandão MLL, Lacerda MLGG, Rocha CABC, Leone de Oliveira SMDV, Parpinelli TC, et al. Fatal Cronobacter sakazakii sequence type 494 meningitis in a newborn, Brazil. Emerg Infect Dis. 2018;24:1948–50. DOIPubMedGoogle Scholar
- Phair K, Pereira SG, Kealey C, Fanning S, Brady DB. Insights into the mechanisms of Cronobacter sakazakii virulence. Microb Pathog. 2022;169:
105643 . DOIPubMedGoogle Scholar - Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricija L, et al. Comparative analysis of genome sequences covering the seven cronobacter species. PLoS One. 2012;7:
e49455 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: August 08, 2023
1These authors contributed equally to this article.
Table of Contents – Volume 29, Number 9—September 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Qingping Wu, Guangdong Provincial Key Laboratory of Microbial Safety and Health, State Key Laboratory of Applied Microbiology, Southern China, Institute of Microbiology, Guangdong Academy of Sciences, Xianlie Zhong Rd 100, Guangzhou City 510070, China
Top