Volume 30, Number 1—January 2024
Research
Disease-Associated Streptococcus pneumoniae Genetic Variation
Figure 2
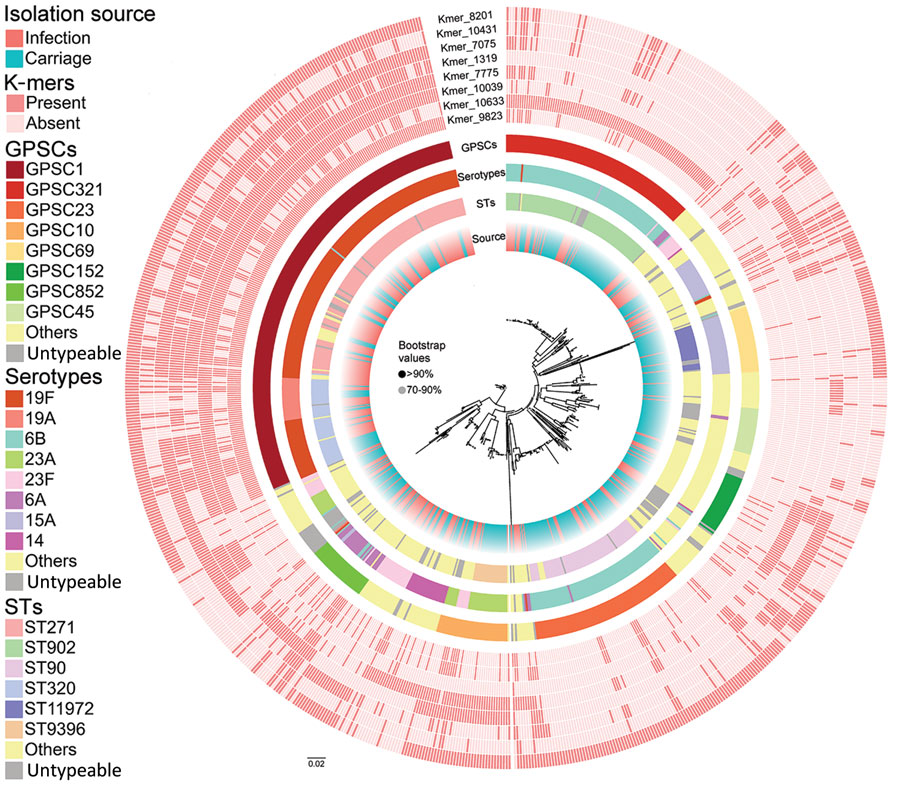
Figure 2. Whole-genome phylogenetic tree showing genetic similarity of 783 Streptococcus pneumoniae isolates in a study of disease-associated Streptococcus pneumoniae genetic variation. The colored strips at the tips of the tree (from inner to outer) represent isolate metadata (source, STs, serotypes, and GPSCs) and infection-associated k-mers found in the final model. GPSC, global pneumococcal sequencing cluster; ST, sequence type.
1These authors contributed equally to this article.
Page created: November 24, 2023
Page updated: December 20, 2023
Page reviewed: December 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.