Volume 30, Number 2—February 2024
Research Letter
SARS-CoV-2 Infection in Beaver Farm, Mongolia, 2021
Abstract
We report an outbreak of COVID-19 in a beaver farm in Mongolia in 2021. Genomic characterization revealed a unique combination of mutations in the SARS-CoV-2 of the infected beavers. Based on these findings, increased surveillance of farmed beavers should be encouraged.
The COVID-19 pandemic that began in 2019 remains uncontained, and fatalities and multiple waves of infection continue to occur worldwide (1). The causative agent, SARS-CoV-2, has been detected in humans and several animal species, including domestic, wild, and laboratory animals (2,3). Because SARS-CoV-2 can be transmitted from humans to animals and back to humans, understanding the dynamics of infection in animals can contribute to the creation of more comprehensive response strategies.
We identified SARS-CoV-2 infection in beavers (Castor fiber) farmed for conservation reasons in Mongolia and report on serologic and whole genome sequence data from this outbreak. The beaver farm, located in the Bayanzurkh district in Ulaanbaatar, Mongolia, reared 32 adults and 16 kits in 2021. They were housed indoors in a large area separated by waist-high walls, with space for multiple animals. One of the 7 employees of the farm had influenza-like symptoms for several days and was diagnosed with COVID-19 on August 6, 2021. On August 9, the beaver farm reported the death of 2 beavers (one 6 months of age and one 2 years of age) after signs of coughing, nasal discharge, rasping on auscultation of the lungs and chest cavity, sluggish movement, and aversion to food. On August 13, research investigators collected nasal swabs, saliva, and 7 tissue samples (lung, kidney, liver, heart, spleen, larynx, and tongue from the 2 dead animals. Researchers also collected nasal swab specimens, saliva, and blood from 7 other beavers with notable clinical signs of coughing and purulent nasal discharge. Follow-up investigation on August 18 or 19 and on September 12 included collection of additional nasal swab specimens, saliva, and blood samples from the same animals as well as from 2 healthy animals (September 12 only).
All samples were transported to a Biosafety Level 3 facility in Ulaanbaatar and were screened by quantitative reverse transcription PCR according to the Peiris protocol (4). The results showed that 46 of 48 specimens from 9 animals with clinical signs, including the 2 dead animals, tested positive for SARS-CoV-2 RNA. Serum was separated from the blood samples by centrifugation (2,000 × g for 10 min) and stored at −20°C until required. The serum samples were then subjected to antibody screening by using a commercial ELISA kit (ID Screen SARS-CoV-2 Double Antigen Multi-species ELISA; Innovative Diagnostics, https://www.innovative-diagnostics.com). Fifteen of 23 samples tested positive and 1 was intermediate, indicating that all animals became antibody positive within 1 month of confirmation of SARS-CoV-2 RNA positivity. One clinically unremarkable beaver tested positive for SARS-CoV-2 antibodies, indicating a possible subclinical infection (Table).
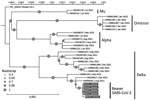
We shipped 5 randomly selected quantitative reverse transcription PCR–confirmed SARS-CoV-2–positive RNA samples to the Animal Production and Health Laboratory (Seibersdorf, Austria), a joint program of the International Atomic Energy Agency and the Food and Agriculture Organization of the United Nations, and subjected them to whole-genome sequencing (Appendix 1; Appendix 2). Based on genotype analysis, all 5 genome sequences were assigned to the B.1.617.2 lineage, commonly referred to as the Delta variant. At the time of sampling, Alpha and Delta variants of SARS-CoV-2 were being identified in humans in Mongolia. The closest related sequences to those we identified in the beavers studied were from human SARS-CoV-2 in Mongolia (GenBank accession nos. ON008302, OM190617, and OM961234) identified during April–September 2021 (Figure). In addition to 4 mutations in the spike region, the sequences shared 7 amino acid substitutions in open reading frame [ORF] 1a, 4 amino acid substitutions in ORF1b, and 1 amino acid substitution in nucleocapsid genes. In the beaver sequences, 4 amino acid substitutions identified were not in the human isolates from Mongolia: S2500F, A3657V in ORF1a and H604Y, T1404M in ORF1b. Although those substitutions have been identified individually in SARS-CoV-2 sequences in GenBank and the GISAID database (https://www.gisaid.org), there are no records of sequences with all 4 mutations.
Several cases of SARS-CoV-2 transmission between humans and animals have already been reported (5–8). An alarming aspect of SARS-CoV-2 infection in animals is that host animals can maintain the virus and contribute to the emergence in humans of new variants that have accumulated multiple mutations (7–10). Indeed, the specific combination of mutations observed in the beavers we studied has not been found in other SARS-CoV-2 sequences in public databases (as of November 2023). This finding suggests that the mutations might have occurred or accumulated after the introduction of the virus into the beaver population. Because the emergence of viruses with mutations not targeted by current SARS-CoV-2 vaccines is a credible possibility, more active surveillance of SARS-CoV-2 infection in animals should be encouraged to identify the appearance of mutated viruses. In intensively farmed animals, species–species and species–humans contact is more frequent than in animals dwelling in other environments, which might increase the risk for zoonotic pathogen transmission (2). Thus, implementing more active surveillance and infection control strategies is critical to disease prevention and containment.
Dr Takemura is a technical expert for the ZODIAC project, Animal Production and Health Laboratory, Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture. His main research interests are in the biology of infectious pathogens.
Acknowledgments
Samples used in this study were those submitted to the State Central Veterinary Laboratory, Mongolia, for emergency diagnosis of SARS-CoV-2. Ethics approval was not required.
Funding came from the VETLAB network initiative of the Joint Food and Agriculture Organization of the United Nations (FAO)/International Atomic Energy Agency (IAEA) Centre through the IAEA Peaceful Uses Initiative Project (“Detection of emerging and re-emerging animal and zoonotic pathogens at the animal-human interface”), funded by the Government of Japan and the United States of America and IAEA ZODIAC Project.
References
- Coronavirus disease (COVID-19) pandemic, World Health Organization web database [cited 2023 Sep 20]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Cui S, Liu Y, Zhao J, Peng X, Lu G, Shi W, et al. An updated review on SARS-CoV-2 infection in animals. Viruses. 2022;14:1527. DOIPubMedGoogle Scholar
- Rao SS, Parthasarathy K., Sounderrajan V, Neelagandan K, Anbazhagan P, Chandramouli V. Susceptibility of SARS Coronavirus-2 infection in domestic and wild animals: a systematic review. 3 Biotech. 2023;13:5.
- World Health Organization Detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by RT-PCR [cited 2023 Nov 1]. https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf
- SARS-CoV-2 in animals situation update, FAO, Rome. [cited 2023 Sep 1]. https://www.fao.org/animal-health/situation-updates/sars-cov-2-in-animals
- Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–7. DOIPubMedGoogle Scholar
- Pickering B, Lung O, Maguire F, Kruczkiewicz P, Kotwa JD, Buchanan T, et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission.[ [Erratum in: Nat Microbiol. 2023:8:188; ]. Nat Microbiol. 2022;7:2011–24. DOIPubMedGoogle Scholar
- Tan CCS, Lam SD, Richard D, Owen CJ, Berchtold D, Orengo C, et al. Transmission of SARS-CoV-2 from humans to animals and potential host adaptation. Nat Commun. 2022;13:2988. DOIPubMedGoogle Scholar
- Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect Dis. 2021;21:18–9. DOIPubMedGoogle Scholar
- Sharun K, Tiwari R, Saied AA, Dhama K. SARS-CoV-2 vaccine for domestic and captive animals: An effort to counter COVID-19 pandemic at the human-animal interface. Vaccine. 2021;39:7119–22. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: January 18, 2024
1These first authors contributed equally to this article.
Table of Contents – Volume 30, Number 2—February 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Taichiro Takemura, Animal Production and Health Laboratory, International Atomic Energy Agency, Friedenstrasse 1, Seibersdorf, Austria
Top