Volume 30, Number 3—March 2024
Research Letter
Source Tracing of Leishmania donovani in Emerging Foci of Visceral Leishmaniasis, Western Nepal
Figure
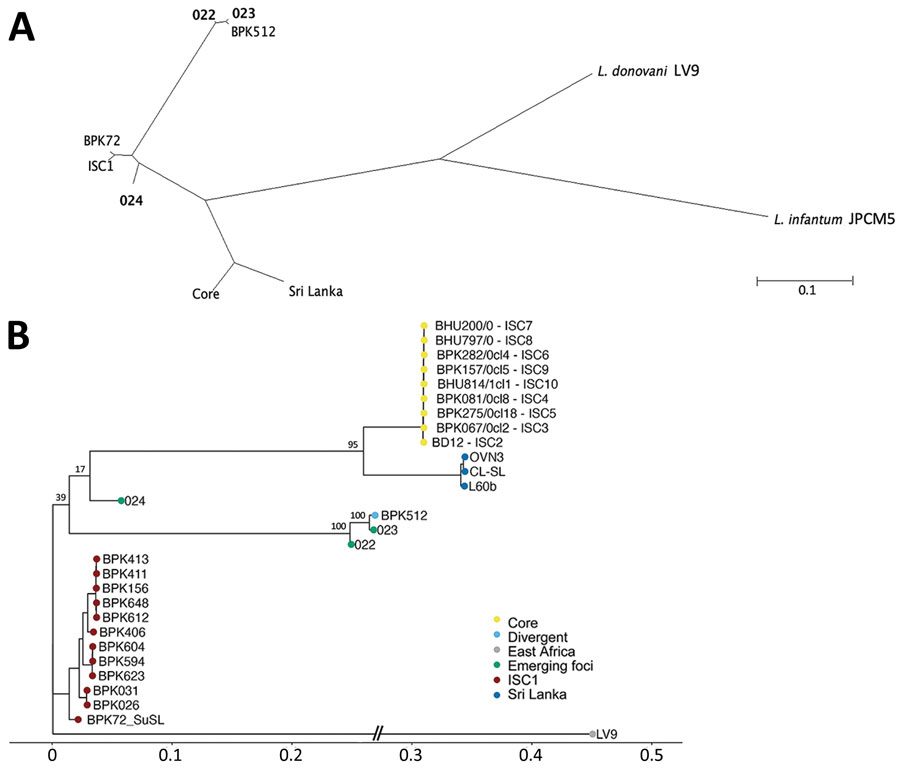
Figure. Phylogenetic analyses of Leishmania donovani from the ISC, including Nepal, and reference sequences. Trees were based on genomewide single-nucleotide polymorphisms using RAxML (8). A) Unrooted phylogenetic network of the L. donovani complex, showing samples representing the emerging foci (bold text). B) Rooted phylogenetic tree of reference strains of L. donovani from the ISC, showing the branching of 3 samples (022, 023, and 024) originating from emerging foci. Important bootstrap values are indicated on the branches. The West-African LV9 strain is included as an outgroup. BPK72_SuSL represents an ISC1 sample analyzed using SureSelect sequencing (Agilent Technologies; https://www.agilent.com), confirming that the branching of the emerging foci is not a result of a technical artifact. Scale bars indicate number of single-nucleotide polymorphism differences. ISC, Indian subcontinent.
References
- Pandey K, Dumre SP, Shah Y, Acharya BK, Khanal L, Pyakurel UR, et al. Forty years (1980-2019) of visceral leishmaniasis in Nepal: trends and elimination challenges. Trans R Soc Trop Med Hyg. 2023;117:460–9. DOIPubMedGoogle Scholar
- Pandey K, Bastola A, Haiyan G, Pyakurel UR, Pandey BD, Dumre SP. Emergence of cutaneous leishmaniasis in Nepal. Trop Med Health. 2021;49:72. DOIPubMedGoogle Scholar
- Domagalska MA, Dujardin JC. Next-generation molecular surveillance of TriTryp diseases. Trends Parasitol. 2020;36:356–67. DOIPubMedGoogle Scholar
- Domagalska MA, Imamura H, Sanders M, Van den Broeck F, Bhattarai NR, Vanaerschot M, et al. Genomes of Leishmania parasites directly sequenced from patients with visceral leishmaniasis in the Indian subcontinent. Rogers MB, editor. PLoS Negl Trop Dis. 2019;13:e0007900.
- Imamura H, Downing T, Van den Broeck F, Sanders MJ, Rijal S, Sundar S, et al. Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. eLife. 2016;5:
e12613 . DOIPubMedGoogle Scholar - Franssen SU, Durrant C, Stark O, Moser B, Downing T, Imamura H, et al. Global genome diversity of the Leishmania donovani complex. eLife. 2020;9:
e51243 . DOIPubMedGoogle Scholar - Zhang WW, Ramasamy G, McCall LI, Haydock A, Ranasinghe S, Abeygunasekara P, et al. Genetic analysis of Leishmania donovani tropism using a naturally attenuated cutaneous strain. PLoS Pathog. 2014;10:
e1004244 . DOIPubMedGoogle Scholar - Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–5. DOIPubMedGoogle Scholar
- Rai K, Bhattarai NR, Vanaerschot M, Imamura H, Gebru G, Khanal B, et al. Single locus genotyping to track Leishmania donovani in the Indian subcontinent: Application in Nepal. PLoS Negl Trop Dis. 2017;11:
e0005420 . DOIPubMedGoogle Scholar - Seblova V, Dujardin JC, Rijal S, Domagalska MA, Volf P. ISC1, a new Leishmania donovani population emerging in the Indian sub-continent: Vector competence of Phlebotomus argentipes. Infect Genet Evol. 2019;76:
104073 . DOIPubMedGoogle Scholar