Volume 30, Number 4—April 2024
Research Letter
Link between Monkeypox Virus Genomes from Museum Specimens and 1965 Zoo Outbreak
Figure
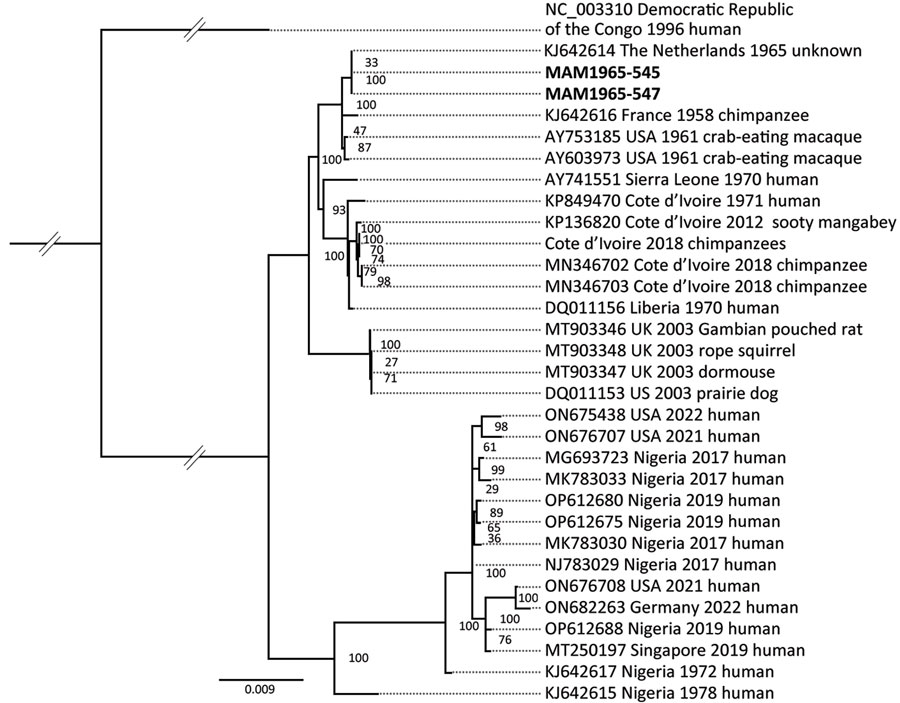
Figure. Maximum-likelihood phylogeny tree showing the close relation between MPXV genomes from museum orangutan samples from Germany (bold text), which fall into clade IIa, to the genome derived from the MPXV zoo outbreak in Rotterdam, the Netherlands, 1965. The phylogeny tree is rooted on the outgroup genome (GenBank accession no. NC_003310) from clade I with the museum orangutan genomes MAM1965–545 and MAM1965–547. The consensus sequences for the ancient sequences are based on a mapping to the Rotterdam genome. The final single-nucleotide polymorphisms alignment length was 138,240 bp. The collapsed node contains genomes from Pan troglodytes verus from Cote d’Ivoire (GenBank accession nos. MN346690, MN346692, MN346694–8, MN346700–1).
1These authors contributed equally to this article.