Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Volume 30, Number 8—August 2024
Research Letter
Rare Case of Echinostoma cinetorchis Infection, South Korea
Figure 2
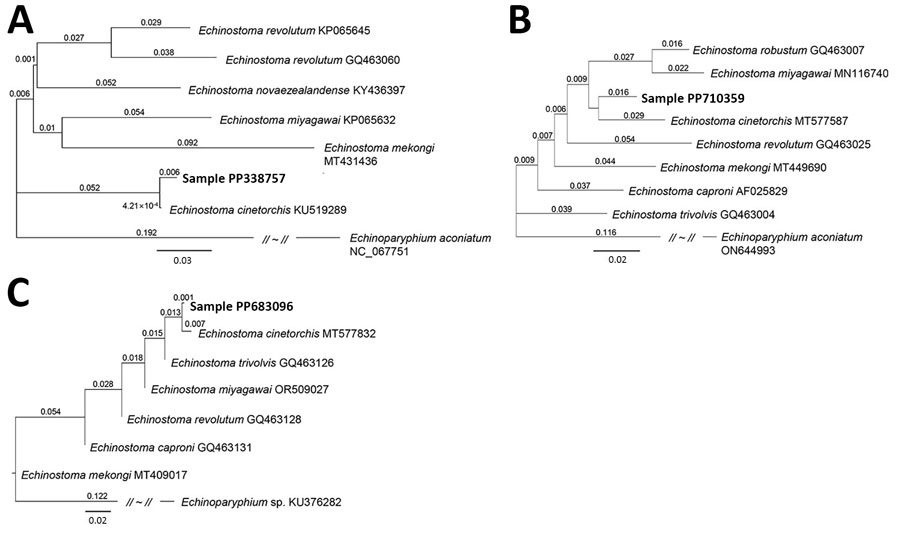
Figure 2. Phylogenetic trees of a worm identified as Echinostoma cinetorchis removed during colonoscopy from a 69-year-old woman in South Korea (red text). Trees were based on nucleotide sequences of the NADH dehydrogenase 1 gene (A), cytochrome c oxidase subunit 1 mitochondrial gene (B), and internal transcribed spacer region (C) of the worm in comparison with various echinostome species deposited in GenBank (accession numbers shown), inferred by the neighbor-joining method (1,000 bootstrap replications) using the Geneious Prime Program 2023.1.2. (Geneious, https://www.geneious.com). Echinoparyphium aconiatum (NADH dehydrogenase 1 and cytochrome c oxidase subunit 1 mitochondrial genes) and Echinoparyphium sp. (internal transcribed spacer) were used as the outgroups. Scale bars indicate evolutionary distance.