Volume 6, Number 5—October 2000
Perspective
Using DNA Microarrays to Study Host-Microbe Interactions
Figure 2
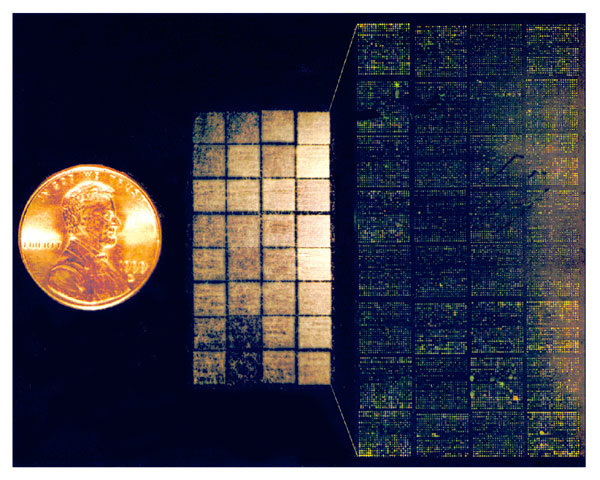
Figure 2. DNA microarray--Lymphochip. (Center) Lymphochip version 8.0, printed on a coated glass microscope slide using a 32-tip printing head, contains 17,856 cDNA clones (overhead illumination) (14). (Left) U.S. penny, for scale. (Right) Scanned image demonstrating differential hybridization of Cy3- and Cy5-labeled cDNA to this microarray. (Illustration by A. Alizadeh, M. Eisen, and P. Brown, Stanford University; and L. Staudt, National Cancer Institute).
References
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–74. DOIPubMedGoogle Scholar
- Carulli JP, Artinger M, Swain PM, Root CD, Chee L, Tulig C, High throughput analysis of differential gene expression. J Cell Biochem Suppl. 1998;30-31:286–96. DOIPubMedGoogle Scholar
- Svanborg C, Godaly G, Hedlund M. Cytokine responses during mucosal infections: role in disease pathogenesis and host defence. Curr Opin Microbiol. 1999;2:99–105. DOIPubMedGoogle Scholar
- Cotter PA, Miller JF. In vivo and ex vivo regulation of bacterial virulence gene expression. Curr Opin Microbiol. 1998;1:17–26. DOIPubMedGoogle Scholar
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. DOIPubMedGoogle Scholar
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–97.PubMedGoogle Scholar
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. DOIPubMedGoogle Scholar
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. DOIPubMedGoogle Scholar
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–6. DOIPubMedGoogle Scholar
- Tao H, Bausch C, Richmond C, Blattner FR, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–40.PubMedGoogle Scholar
- Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–35. DOIPubMedGoogle Scholar
- Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T, Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 1998;58:5009–13.PubMedGoogle Scholar
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96:9212–7. DOIPubMedGoogle Scholar
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. DOIPubMedGoogle Scholar
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. DOIPubMedGoogle Scholar
- Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci U S A. 1999;96:6745–50. DOIPubMedGoogle Scholar
- Wang K, Gan L, Jeffery E, Gayle M, Gown AM, Skelly M, Monitoring gene expression profile changes in ovarian carcinomas using cDNA microarray. Gene. 1999;229:101–8. DOIPubMedGoogle Scholar
- Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–301. DOIPubMedGoogle Scholar
- Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–44. DOIPubMedGoogle Scholar
- Southern E, Mir K, Shchepinov M. Molecular interactions on microarrays. Nat Genet. 1999;21:5–9. DOIPubMedGoogle Scholar
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–7. DOIPubMedGoogle Scholar
- Watson A, Mazumder A, Stewart M, Balasubramanian S. Technology for microarray analysis of gene expression. Curr Opin Biotechnol. 1998;9:609–14. DOIPubMedGoogle Scholar
- Fodor SP, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–6. DOIPubMedGoogle Scholar
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–80. DOIPubMedGoogle Scholar
- Cheung VG, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Making and reading microarrays. Nat Genet. 1999;21:15–9. DOIPubMedGoogle Scholar
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21:10–4. DOIPubMedGoogle Scholar
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–4. DOIPubMedGoogle Scholar
- de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–8. DOIPubMedGoogle Scholar
- Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci U S A. 1999;96:12833–8. DOIPubMedGoogle Scholar
- Talaat AM, Hunter P, Johnston SA. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat Biotechnol. 2000;18:679–82. DOIPubMedGoogle Scholar
- Cohen P, Bouaboula M, Bellis M, Baron V, Jbilo O, Poinot-Chazel C, Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J Biol Chem. 2000;275:11181–90. DOIPubMedGoogle Scholar
- Eckmann L, Smith JR, Housley MP, Dwinell MB, Kagnoff MF. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria salmonella. J Biol Chem. 2000;275:14084–94. DOIPubMedGoogle Scholar
- Rosenberger CM, Scott MG, Gold MR, Hancock RE, Finlay BB. Salmonella Typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J Immunol. 2000;164:5894–904.PubMedGoogle Scholar
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–7. DOIPubMedGoogle Scholar
- Geiss GK, Bumgarner RE, An MC, Agy MB, van 't Wout AB, Hammersmark E, Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000;266:8–16. DOIPubMedGoogle Scholar
- Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:14470–5. DOIPubMedGoogle Scholar
- Bassett DE Jr, Eisen MB, Boguski MS. Gene expression informatics--it's all in your mine. Nat Genet. 1999;21:51–5. DOIPubMedGoogle Scholar
- Khan J, Bittner ML, Chen Y, Meltzer PS, Trent JM. DNA microarray technology: the anticipated impact on the study of human disease. Biochim Biophys Acta. 1999;1423:M17–28.PubMedGoogle Scholar
- Brazma A, Robinson A, Cameron G, Ashburner M. One-stop shop for microarray data. Nature. 2000;403:699–700. DOIPubMedGoogle Scholar
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–12. DOIPubMedGoogle Scholar
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. DOIPubMedGoogle Scholar
- Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. J Comput Biol. 1999;6:281–97. DOIPubMedGoogle Scholar
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–35. DOIPubMedGoogle Scholar
- Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci U S A. 2000;97:262–7. DOIPubMedGoogle Scholar
- Gaasterland T, Bekiranov S. Making the most of microarray data. Nat Genet. 2000;24:204–6. DOIPubMedGoogle Scholar
- Berry MJA, Linoff G. Data mining techniques for marketing, sales and customer support. New York: John Wiley and Sons;1997.
- Weaver DC, Workman CT, Stormo GD. Modeling regulatory networks with weight matrices. In: Altman RB, Lauderdale K, Dunker AK, Hunter L, Klein TE, editors. Biocomputing '99: Proceedings of the Pacific Symposium. River Edge (NJ): World Scientific Press;1999:112-23.
- Chen T, He HL, Church GM. Modeling gene expression with differential equations. In: Altman RB, Lauderdale K, Dunker AK, Hunter L, Klein TE, editors. Biocomputing '99: Proceedings of the Pacific Symposium. River Edge (NJ): World Scientific Press; 1999:29-40.
- Davidson GS, Hendrickson B, Johnson DK, Meyers CE, Wylie BN. Knowledge mining with VxInsight: discovery through interaction. Journal of Intelligent Information Systems. Integrating Artificial Intelligence and Database Technologies. 1998;11:259–85.
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–41. DOIPubMedGoogle Scholar
- Ramakrishna R, Srinivasan R. Gene identification in bacterial and organellar genomes using GeneScan. Comput Chem. 1999;23:165–74. DOIPubMedGoogle Scholar
- Hayes WS, Borodovsky M. How to interpret an anonymous bacterial genome: machine learning approach to gene identification. Genome Res. 1998;8:1154–71.PubMedGoogle Scholar
- Audic S, Claverie JM. Self-identification of protein-coding regions in microbial genomes. Proc Natl Acad Sci U S A. 1998;95:10026–31. DOIPubMedGoogle Scholar
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–15. DOIPubMedGoogle Scholar
- Borodovsky M, McIninch J. Recognition of genes in DNA sequence with ambiguities. Biosystems. 1993;30:161–71. DOIPubMedGoogle Scholar
- Rozen S, Skaletsky HJ. Primer3; 1996, 1997, 1998. Cambridge (MA): Whitehead Institute. Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html.
- Hayward RE, Derisi JL, Alfadhli S, Kaslow DC, Brown PO, Rathod PK. Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol Microbiol. 2000;35:6–14. DOIPubMedGoogle Scholar
- Koonin EV, Mushegian AR, Galperin MY, Walker DR. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol Microbiol. 1997;25:619–37. DOIPubMedGoogle Scholar
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–6. DOIPubMedGoogle Scholar
- Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan JS, DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–66.PubMedGoogle Scholar
- Mekalanos JJ. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7.PubMedGoogle Scholar
- Mangan JA, Monahan IM, Wilson MA, Schnappinger D, Schoolnik GK, Butcher PD. The expression profile of Mycobacterium tuberculosis infecting the human monocytic cell line THP-1 using whole genome microarray analysis. Nat Genet. 1999;23:61. DOIGoogle Scholar
- Chiang SL, Mekalanos JJ, Holden DW. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol. 1999;53:129–54. DOIPubMedGoogle Scholar
- Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–61. DOIPubMedGoogle Scholar
- Sokolovic Z, Schuller S, Bohne J, Baur A, Rdest U, Dickneite C, Differences in virulence and in expression of PrfA and PrfA-regulated virulence genes of Listeria monocytogenes strains belonging to serogroup 4. Infect Immun. 1996;64:4008–19.PubMedGoogle Scholar
- Bohne J, Kestler H, Uebele C, Sokolovic Z, Goebel W. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol Microbiol. 1996;20:1189–98. DOIPubMedGoogle Scholar
- Debouck C, Goodfellow PN. DNA microarrays in drug discovery and development. Nat Genet. 1999;21:48–50. DOIPubMedGoogle Scholar
- Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–8. DOIPubMedGoogle Scholar
- Troesch A, Nguyen H, Miyada CG, Desvarenne S, Gingeras TR, Kaplan PM, Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J Clin Microbiol. 1999;37:49–55.PubMedGoogle Scholar
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–6. DOIPubMedGoogle Scholar
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–3. DOIPubMedGoogle Scholar
- Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A. 1999;96:13264–9. DOIPubMedGoogle Scholar
- Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–69.PubMedGoogle Scholar
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. DOIPubMedGoogle Scholar
- Corbeil J, Sheeter D, Rought S, Du P, Ferguson M, Masys DR, Magnitude and specificity of temporal gene expression during HIV-1 infection of a CD4+ T cell. Nat Genet. 1999;23:39–40. DOIGoogle Scholar
- Xiang C, Young H, Alterson H, Reynolds D, Bittner M, Chen Y, Comparison of cellular gene expression in Ebola-Zaire and Ebola-Reston virus-infected primary human monocytes. Nat Genet. 1999;23:82. DOIGoogle Scholar
- Nuwaysir EF, Bittner M, Trent J, Barrett JC, Afshari CA. Microarrays and toxicology: the advent of toxicogenomics. Mol Carcinog. 1999;24:153–9. DOIPubMedGoogle Scholar
- Roebuck KA, Carpenter LR, Lakshminarayanan V, Page SM, Moy JN, Thomas LL. Stimulus-specific regulation of chemokine expression involves differential activation of the redox-responsive transcription factors AP-1 and NF-kappaB. J Leukoc Biol. 1999;65:291–8.PubMedGoogle Scholar
- Lo D, Feng L, Li L, Carson MJ, Crowley M, Pauza M, Integrating innate and adaptive immunity in the whole animal. Immunol Rev. 1999;169:225–39. DOIPubMedGoogle Scholar
Page created: December 17, 2010
Page updated: December 17, 2010
Page reviewed: December 17, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.