Volume 10, Number 6—June 2004
Research
Candida parapsilosis Characterization in an Outbreak Setting
Abstract
Candida parapsilosis is an important non-albicans species which infects hospitalized patients. No studies have correlated outbreak infections of C. parapsilosis with multiple virulence factors. We used DNA fingerprinting to determine genetic variability among isolates from a C. parapsilosis outbreak and from our clinical database. We compared phenotypic markers of pathogenesis, including adherence, biofilm formation, and protein secretion (secretory aspartic protease [SAP] and phospholipase). Adherence was measured as colony counts on silicone elastomer disks immersed in agar. Biofilms formed on disks were quantified by dry weight. SAP expression was measured by hydrolysis of bovine albumin; a colorimetric assay was used to quantitate phospholipase. DNA fingerprinting indicated that the outbreak isolates were clonal and genetically distinct from our database. Biofilm expression by the outbreak clone was greater than that of sporadic isolates (p ≤ 0.0005). Adherence and protein secretion did not correlate with strain pathogenicity. These results suggest that biofilm production plays a role in C. parapsilosis outbreaks.
The yeast Candida is the fourth most common cause of hospital-related bloodstream infections (1). Forty percent of patients who have had Candida isolated from their intravenous catheters have underlying fungemia (2), and the case-fatality rate for catheter-related candidemia approaches 40% (3).
Although C. albicans is the most commonly isolated yeast, other species are found with increasing frequency, including C. parapsilosis (4). C. parapsilosis particularly affects critically ill neonates and surgical intensive care unit (ICU) patients (5,6), likely because of its association with parenteral nutrition and central lines (7,8). The affinity of C. parapsilosis for foreign material is shown by infections related to peritoneal dialysis catheters (9) and prosthetic heart valves (10), and this characteristic may be important in infections of cancer patients with indwelling access devices (11). C. parapsilosis is increasingly responsible for hospital outbreaks, and the hands of healthcare workers may be the predominant environmental source (12).
Our understanding of fungal virulence factors is limited. The surface adherence capacity of Candida is likely one such factor, possibly linked to its subsequent ability to form biofilms (13). Clinically obtained C. albicans isolates form biofilms (14), which may be important for sustaining infection. Adhesion (15) and biofilm formation (14) may be especially important for C. parapsilosis, since indwelling devices appear to be the predominant route of infection (8,11). Total parenteral nutrition (TPN) solutions may promote C. parapsilosis adhesion and growth (16). Recently, biofilm-forming potential was cited as a reason that patients with C. parapsilosis-infected catheters should have the device removed (17).
Fungi secrete enzymes integral to pathogenesis. Phospholipases (e.g., phospholipase B) and proteases (e.g., secreted aspartyl proteases [SAPs]) are two of the best-characterized. Although phospholipase B expression has been well studied in C. albicans (18), the relationship between C. parapsilosis virulence and phospholipase phenotype is unclear. The role of SAP and pathogenesis is similarly unclear (19).
We characterized genetic and phenotypic characteristics of isolates from a C. parapsilosis outbreak that occurred in a Mississippi community hospital (20) and compared these characteristics with those of isolates obtained from persons with sporadic infections at our tertiary hospital. We performed molecular characterization, comparing C. parapsilosis isolates involved in the outbreak with those from our own clinical collection. We then compared adhesion ability, biofilm production, and secretion of SAP and phospholipase B of the outbreak isolates and our clinical strains.
Organisms
Outbreak isolates of C. parapsilosis were obtained at a Mississippi hospital from April through October 2001. Epidemiologic details regarding the organisms and patients have been published elsewhere (21). We examined five invasive strains (defined as obtained from blood or catheter cultures: 165, 167, 173, 177, 179), and three environmental isolates (from healthcare workers’ hands; 313, 317, and 385). Isolates from sporadic infections were from the culture collection at the Center for Medical Mycology, University Hospitals of Cleveland (University Hospitals). C. parapsilosis strain P/A71 was obtained from sputum, P92 from blood, and C. albicans M61 from an intravascular line (19). The site of isolation of other strains is indicated in the figures. Speciation was performed by using germ tube tests and API20C-AUX methods. Organism propagation has been described previously (21). All specimens were stored and used without patient identifiers, to maintain confidentiality.
DNA Fingerprinting
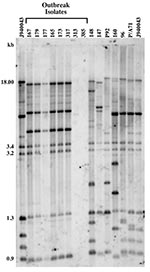
Isolates were analyzed by Southern blot hybridization using the complex DNA fingerprinting probe Cp3-13 (22), according to published methods (23). Genomic DNA was extracted from cells according to the protocol described by Scherer and Stevens (24). Three micrograms of DNA preparation were digested with a combination of EcoRI and SalI (4 U each per microgram of DNA) for 16 h at 37°C, then underwent electrophoresis at 60 V in a 0.7% agarose gel. C. parapsilosis strain J940043 was used as a reference, and its DNA was run in the first and last lanes of the fingerprinting gel. The DNA was transferred from the gel to a nylon Hybond N+ membrane (Amersham, Piscataway, NJ) by capillary blotting, prehybridized with sheared salmon sperm DNA, hybridized overnight with [32P]dCTP-labeled Cp3-13 probe, and viewed by autoradiogram.
Computer-Assisted Cluster Analysis
The autoradiogram image was digitized, unwarped, and straightened, by using the DENDRON software database (25). Processed hybridization patterns were scanned to identify and link common bands. Patterns underwent pairwise comparison: the similarity coefficient (SAB) between the patterns of every pair of isolates A and B was computed according to the formula:
SAB=2E/(2E + a + b)
where E is the number of bands common to both strains, a is the number of bands unique to strain A, and b is the number of bands unique to strain B (22). The SAB ranges from 0.0 (no common bands) to 1.0 (identical match of all bands). Dendrograms based on SAB values were generated by the unweighted pair-group method with arithmetic average (UPGMA) (26); values of 0.07 were considered the threshold for group association (27).
Adherence Assays
Adherence of C. parapsilosis isolates to silicon elastomer (SE) disks was measured by using a modification of earlier methods (28); SE was obtained from Cardiovascular Instrument Corp. (Wakefield, MA) and prepared as described (14). Standardized suspensions of 50 to 200 cells/mL were used for injection into SE disks. After injection, disks were washed in phosphate-buffered saline (PBS) to remove nonadherent cells and placed in wells of 12-well tissue culture plates (Becton Dickinson, Franklin Lakes, NJ). Two milliliters of warm (55°C) liquid SD agar was added per well to completely cover the SE disks and allowed to solidify. Plates were incubated overnight (37°C), and colonies adhering per disk were counted by using a dissecting microscope.
Biofilm Formation and Quantitation
C. parapsilosis biofilms were formed on SE disks as described previously (14). Control disks were handled identically, except that no blastospores were added. Biofilm quantitation was performed as described (21) with dry weight measurements. Dry weight measured total biofilm mass including fungal cells and extracellular matrix.
SAP Assays
Previous authors have described methods of evaluating the ability of Candida SAP to degrade bovine serum albumen (BSA) from cells grown in SAP expression media (29). We grew C. parapsilosis isolates in yeast nitrogen base (YNB) because we could detect SAP activity using YNB, the use of expression medium resulted in contaminating BSA bands, and our assay utilized the same medium used for examining biofilm formation. To confirm relevance of our findings to those of previous studies, we examined SAP expression of organisms grown in the expression medium and found similar results (data not shown).
After overnight growth, Candida cell suspensions were centrifuged (6000 × g for 8 min), and the supernatant was collected, then concentrated by using a Centricon 10,000 NMWL filter centrifuge (Millipore Corp., Billerica, MA). Supernatant protein (500 ng) was incubated at 37°C for 15 min with 0.4 mL of 1% BSA (wt/vol, in 0.1 mol/L citrate buffer, pH 3.2). After incubation, 10 μL sodium dodecyl sulfate (SDS) sample buffer and 7 μL of reducing agent were added to 40 μL of each mixture, and the proteins solubilized by boiling (10 min). Ten microliters of sample was separated by SDS–polyacrylamide gel electrophoresis (PAGE), and the protein bands were visualized by silver staining (SilverXpress Staining Kit, Invitrogen Corp., Carlsbad, CA). The appearance of a 20-kDa band was indicative of SAP activity. Quantitation of this band was determined by using QuantOne software v4.3.0 (BioRad Laboratories, Hercules, CA). Control experiments were performed by adding either no supernatant (100 μL of sodium citrate buffer instead) or supernatant mixed with protease inhibitor cocktail (Sigma Chemical Co., St. Louis, MO); 10 μL/mL of supernatant). Protein estimations were performed by using the BioRad Dc kit (BioRad Laboratories) and BSA as standard.
Phospholipase Assays
A colorimetric assay for free fatty acid (FFA) was used to assess phospholipase activity (30). The incubation mixture for phospholipase (acylhydrolase) activity consisted of 200 μM dipalmitoyl (C16:0) phosphatidylcholine and 200 μmol/L L-palmitoylcarnitine in 0.1% (v/v) Triton X-100. Concentrated culture supernatant was added (100 μg of total protein), and the mixture made up to a final volume of 0.25 mL with 0.1 mol/L of sodium citrate, pH 4.0. Reactions were incubated at 37°C for 1 h, then stopped by adding chloroform/methanol (1:2, v/v). The reaction products were extracted (31), evaporated to dryness under nitrogen, and taken up in 50 μL of 0.1% (vol/vol) Triton X-100. The relative level of free fatty acids in each sample was determined by using an acyl-CoA-oxidase system assay kit (Roche Molecular Biochemicals, Indianapolis, IN).
Statistical Analysis
Adherence and biofilm experiments were performed in quadruplicate and on separate days. Results for different isolates were normalized to C. parapsilosis strain 167 to facilitate meaningful comparisons across multiple experiments (14). Phospholipase and SAP assays were performed at least twice; representative results are shown. Statistical analysis was performed by using StatView v5.0.1 software (SAS Institute, Cary, NC); p values < 0.05 were considered significant.
DNA Fingerprinting Analysis
Isolate relatedness was investigated by using the complex DNA fingerprinting probe Cp3-13 (22). We examined both outbreak strains and our independent University Hospitals’ isolates to characterize the relatedness of a range of clinical C. parapsilosis strains. As shown in Figure 1, the five invasive strains and one of the three environmental isolates generated identical patterns. The two remaining strains (313 and 385) were hand isolates. The fingerprinting pattern of strain 313 was limited to weak bands, while none were obtained for 385. The patterns of the outbreak isolates were also distinct from those of the University Hospitals’ isolates.
Relatedness among outbreak and University Hospitals’ isolates was further assessed through cluster analysis (Figure 2). The six outbreak isolates with an identical fingerprinting pattern had an SAB value close to 1, whereas the dendrogram nodes linking the remaining isolates to each other had an SAB value <0.7, the threshold for relatedness (27). These analyses showed that the six outbreak isolates were identical and belonged to group I strains (22). The remaining isolates appeared moderately related to unrelated at the genetic level, and therefore were non–group I strains. Previous studies have shown that Cp3-13 fingerprinting patterns made up of a few weak bands typically belong to groups II or III, representing a minority of C. parapsilosis clinical isolates (22,32). However, internally transcribed spacer region sequencing of strain 313 indicates that it, in fact, belongs to group 1 (D. Warnock, pers. comm.). Alternately, this finding may suggest past genetic exchanges between group I and non–group I strains. The dendrogram also shows that, in addition to being unrelated to the outbreak isolates, the University Hospitals’ strains are not related to one another but represent sporadic cases.
Adherence
Adherence to substrate, whether natural (endothelium) or artificial (catheter material), is likely the first step in Candida pathogenesis (33). As shown in Figure 3, the adherence abilities of C. parapsilosis isolates vary widely. Adherence was the same for outbreak isolates of the same clone (167, 165, 173, 317; other clonal isolates were excluded for clarity), regardless of the site of isolation. Adherence was significantly higher than for the two unrelated hand isolates (313 and 385; p < 0.001). No relationship was found between the outbreak isolates and University Hospitals’ isolates, although the latter exhibited higher values than strain 313 and 385. Among University Hospitals’ isolates, no relationship was found between infection site and adherence.
Biofilm Production
We first determined the ability of the C. parapsilosis outbreak isolates to form biofilm. All isolates of the same clone (strains 167, 165, 177, 179, 173, and 317) showed a similar pattern of biofilm formation by dry weight (Figure 4A), which suggests that biofilm formation by these isolates is consistent and not site-induced. Except for the unrelated environmental strains 313 and 385, biofilm formation was not significantly different for the various outbreak isolates (p > 0.05, when compared with strain 167). However, noninvasive strains 313 and 385 produced significantly less biofilm when measured by dry weight (compared with strain 167; p < 0.001 for 385 and p = 0.001 for 313).
Figure 4B shows a comparison of biofilm formation by the C. parapsilosis outbreak clone with isolates obtained from our University Hospitals’ collection, including specimens from different body sites. The outbreak strain 167 produced more biofilm than the University Hospitals’ isolates (p < 0.0005 for comparison of dry weight values of 167 vs. all others), which indicates that outbreak clonal isolates had a higher ability to form biofilms.
For both sets of isolates, biofilm production was examined when TPN solution was substituted for YNB medium because TPN promotes C. parapsilosis growth (16). TPN increased the dry weight of biofilms formed by the clonal strain (167) by up to 40% (p = 0.008). However, the pattern of results across strains was similar to YNB-based method (not shown).
SAP Assays
We measured SAP production by assaying the ability of C. parapsilosis supernatant to hydrolyze BSA (29). A representative SDS-PAGE gel is shown in Figure 5A. The appearance of specific digestion products was noted when culture supernatant was added to BSA. Specifically, we observed the presence of a 20-kDa product, which did not appear when the reaction was carried out in presence of a protease inhibitor cocktail (Figure 5A, lanes marked “+”). This indicated that the 20-kDa band was a specific by-product of supernatant protease activity, present in all supernatants (except for strains M61 and 313). Analysis of the protease activity of culture supernatants was performed by densitometric scanning of the 20-kDa band. As seen in Figure 5B, the intensity of the 20-kDa product varied greatly between strains and within the outbreak clonal isolates, and no consistent pattern of protease activity was evident. These results were confirmed in multiple experiments.
Phospholipase Assays
We determined the phospholipase activity of supernatants obtained from cultures of the different C. parapsilosis isolates, using a colorimetric assay (30). For comparative purposes, we included the phospholipase activity of C. albicans strain M61, since C. albicans is a known phospholipase producer. Although phospholipase activity varied among strains (Figure 6), no consistent differences were observed between sources (outbreak vs. University Hospitals), sites (e.g., blood vs. other), or clonality of isolates.
We studied a nosocomial outbreak of C. parapsilosis (20) and compared outbreak isolates with ones obtained from sporadic infections at our facility. Genetic analysis showed the invasive outbreak isolates (defined as being cultured from blood or catheter (14,34), as well as at least one hand isolate, were from the same clone. This finding agrees with results of previous epidemiologic studies of C. parapsilosis infections, which found predominant clonality (10,35). Since these clones were the same as environmental isolates, the outbreaks appear to have a nosocomial environmental origin. This conclusion also seems to be the case in the Mississippi outbreak. In contrast, the University Hospitals’ strains were unrelated, indicating sporadic infection. Our results support assertions that molecular analysis is useful in investigating C. parapsilosis outbreaks (36).
Adhesion is likely a critical first step in yeast pathogenesis (33). This property might be expected for C. parapsilosis, which is thought to be acquired from exogenous sources, subsequently adheres to indwelling devices, and finally invades the host. However, our results agree with those of DeBernardis (37), who found no difference in adhesion between invasive and skin isolates of C. albicans. Other studies on C. parapsilosis adherence, in fact, showed an inverse relationship between invasiveness and adherence (38).
The clonal outbreak isolates produced more biofilm than either the unrelated environmental strains, or the University Hospitals’ specimens. These results suggest that biofilm formation is an important component of an outbreak strain’s ability to cause infection. Although previous studies of C. albicans (14) and C. parapsilosis (16,39) have suggested that bloodstream isolates produce more biofilm, further in vivo and clinical studies are needed to confirm these findings. If increased production of biofilm by outbreak isolates can be confirmed, this finding may point to a strategy for determining the significance of C. parapsilosis clinical isolates. Finally, our results confirm earlier work, suggesting that TPN promotes the development of C. parapsilosis biofilms (39).
Although secreted enzymes are likely virulence factors for all yeast species, patterns of expression vary across species. This variation may be exemplified by SAP phenotypes. Although SAPs may play a role in C. albicans adhesion (40), they are not important for the primary mode of invasion (through gut mucosa) because knockout strains do not display attenuated virulence (19). However, SAP may be important for pathogenesis at other sites or stages of infection. Regarding C. parapsilosis, DeBernardis et al. (37) found an inverse relationship with invasiveness. Our findings do not support the concept that SAP expression is a critical virulence factor for C. parapsilosis. The variability of our results even across a single clone also raises a larger question of the appropriateness of characterizing SAP expression as a stable experimental pathogenic factor in C. parapsilosis. Although previous work suggested that SAP expression is stable for a given isolate (41), wide variations exist in relevant mRNA production over fairly short periods (42).
Phospholipases are important virulence factors for C. albicans (18) but have not been well studied in C. parapsilosis. Although the isolates examined in this article produced phospholipase, no correlation was found between phospholipase activity and site of infection or other virulence factors. Although one article described phospholipase B and protease activity in a few strains of C. parapsilosis (43), ours is the first study to conduct a more detailed examination of phospholipase behavior in this species.
Our results also show no apparent correlations across the multiple putative virulence factors studied. This result agrees with Branchini’s examination of genotypic variation and slime production in C. parapsilosis (44). Other studies that have reported correlation in expression of multiple virulence factors have been for C. albicans (45), of uncertain relevance to C. parapsilosis.
Although we did not find significant associations between most of the virulence factors and clinical pathologic changes, these results do not mean that these factors are unimportant. Rather, the results suggest that they are not critical to clinical outbreaks. Since all isolates expressed some degree of adhesion (except for hand isolates 313 and 385), SAP, and phospholipase B activity, these phenotypes may be necessary but not sufficient prerequisites for infection. Our results highlight the importance of using outbreak isolates (rather than those from sporadic infections or laboratory stocks) in studies of Candida virulence. Definitive analysis of the role of virulence factors will require further genetic analysis and in vivo models examining the behavior of knockout mutants of C. parapsilosis.
This study has some limitations. First, the number of isolates is small. Given the nature of C. parapsilosis infections and outbreaks, a higher N could not be expected, and the outbreak is especially well-characterized (20). Previous work has drawn conclusions from as few as one isolate or from less-characterized groups of isolates (16). Second, genetic analysis of C. parapsilosis that uses Cp3-13 fingerprinting may have limitations. Third, our SAP assay is unable to determine the relative contributions of different members of the gene family, of which there are at least 10 (47). Such analysis is beyond the scope of this paper. Fourth, similar limitations exist regarding characterization of phospholipase activity, as Candida expresses multiple phospholipases (18). Phospholipase expression varies with environmental conditions (48); however, we performed experiments under standardized conditions, physiologic temperature, and using glucose containing solutions (14).
In conclusion, the genotypic pattern of this C. parapsilosis outbreak suggests a clonal outbreak, likely arising from an environmental source and distinct from sporadic infection. The outbreak clone produced more biofilm than all other strains. No clear relationships were apparent for the other putative virulence factors, which suggests that they are not critical to outbreak behavior. Further genetic and in vivo studies are required to confirm these findings. Future analysis of virulence mechanisms likely needs to use outbreak strains, as well as taking into account the interplay of organism, host, and environment.
Acknowledgments
We thank T. George, N. Isham, and M. Balkis for technical assistance and K. Smith for secretarial assistance.
Funding was provided by the National Institutes of Health (Infectious Disease/Geographic Medicine Training grant no. AI07024, D.M.K.; grants no. AI35097-03, R01-DE13992, and R01-DE13932-01A1, M.A.G.), the American Heart Association (Scientist Development Grant no. 0335313N, P.K.M.), the Dermatology Foundation (Research Fellowship, J.C.), and the Center for AIDS Research at Case Western Reserve University (grant no. AI-36219).
References
- Banerjee SN, Emori TG, Culver DH, Gaynes RP, Jarvis WR, Horan T, Secular trends in nosocomial primary bloodstream infections in the United States, 1980–1989. National Nosocomial Infections Surveillance System. Am J Med. 1991;91:86S–9S. DOIPubMedGoogle Scholar
- Anaissie EJ, Rex JH, Uzun O, Vartivarian S. Predictors of adverse outcome in cancer patients with candidemia. Am J Med. 1998;104:238–45. DOIPubMedGoogle Scholar
- Nguyen MH, Peacock JE, Tanner DC, Morris AJ, Nguyen ML, Snydman DR, Therapeutic approaches in patients with candidemia: evaluation in a multicenter prospective observational study. Arch Intern Med. 1995;155:2429–35. DOIPubMedGoogle Scholar
- Safdar A, Perlin DS, Armstrong D. Hematogenous infections due to Candida parapsilosis: changing trends in fungemic patients at a comprehensive cancer center during the last four decades. Diagn Microbiol Infect Dis. 2002;44:11–6. DOIPubMedGoogle Scholar
- Levy I, Rubin LG, Vasishtha S, Tucci V, Sood SK. Emergence of Candida parapsilosis as the predominant species causing candidemia in children. Clin Infect Dis. 1998;26:1086–8. DOIPubMedGoogle Scholar
- Kao AS, Brandt ME, Pruitt WR, Conn LA, Perkins BA, Stephens DS, The epidemiology of candidemia in two United States cities: results of a population-based active surveillance. Clin Infect Dis. 1999;29:1164–70. DOIPubMedGoogle Scholar
- Saiman L, Ludington E, Pfaller M, Rangel-Frausto S, Wiblin RT, Dawson J, ; National Epidemiology of Mycosis Survey Study Group. Risk factors for candidemia in Neonatal Intensive Care Unit patients. Pediatr Infect Dis J. 2002;19:319–24. DOIPubMedGoogle Scholar
- Weems JJ Jr, Chamberland ME, Ward J, Willy M, Padhye AA, Solomon SL. Candida parapsilosis fungemia associated with parenteral nutrition and contaminated blood pressure transducers. J Clin Microbiol. 1987;25:1029–32.PubMedGoogle Scholar
- Wong PN, Mak SK, Lo KY, Tong GM, Wong AK. A retrospective study of seven cases of Candida parapsilosis peritonitis in CAPD patients: the therapeutic implications. Perit Dial Int. 2000;20:76–9.PubMedGoogle Scholar
- Diekema DJ, Messer SA, Hollis RJ, Wenzel RP, Pfaller MA. An outbreak of Candida parapsilosis prosthetic valve endocarditis. Diagn Microbiol Infect Dis. 1997;29:147–53. DOIPubMedGoogle Scholar
- Levin AS, Costa SF, Mussi NS, Basso M, Sinto SI, Machado C, Candida parapsilosis fungemia associated with implantable and semi-implantable central venous catheters and the hands of healthcare workers. Diagn Microbiol Infect Dis. 1998;30:243–9. DOIPubMedGoogle Scholar
- Lupetti A, Tavanti A, Davini P, Ghelardi E, Corsini V, Merusi I, Horizontal transmission of Candida parapsilosis candidemia in a neonatal intensive care unit. J Clin Microbiol. 2002;40:2363–9. DOIPubMedGoogle Scholar
- Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. Biofilm formation by Candida dubliniensis. J Clin Microbiol. 2001;39:3234–40. DOIPubMedGoogle Scholar
- Kuhn DM, Chandra J, Mukherjee PK, Ghannoum MA. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun. 2002;70:887–98. DOIPubMedGoogle Scholar
- Weems JJ Jr. Candida parapsilosis: epidemiology, pathogenicity, clinical manifestations, and antimicrobial susceptibility. Clin Infect Dis. 1992;14:756–66.PubMedGoogle Scholar
- Pfaller MA, Messer SA, Hollis RJ. Variations in DNA subtype, antifungal susceptibility, and slime production among clinical isolates of Candida parapsilosis. Diagn Microbiol Infect Dis. 1995;21:9–14. DOIPubMedGoogle Scholar
- Pfaller MA. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin Infect Dis. 1996;22(Suppl 2):S89–94.PubMedGoogle Scholar
- Ghannoum MA. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev. 2000;13:122–43. DOIPubMedGoogle Scholar
- Kretschmar M, Felk A, Staib P, Schaller M, Hess D, Callapina M, Individual acid aspartic proteinases (SAPs) 1–6 of Candida albicans are not essential for invasion and colonization of the gastrointestinal tract in mice. Microb Pathog. 2002;32:61–70. DOIPubMedGoogle Scholar
- Clark TA, Morgan J, Brandt M, Lott T, Slavinski S, Taylor S, Abstracts of the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC), San Diego, California, 2002 Sep 27–30. Washington: American Society for Microbiology; 2002.
- Chandra J, Mukherjee PK, Leidich SD, Faddoul FF, Hoyer LL, Douglas LJ, Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J Dent Res. 2001;80:903–8. DOIPubMedGoogle Scholar
- Enger L, Joly S, Pujol C, Simonson P, Pfaller M, Soll DR. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J Clin Microbiol. 2001;39:658–69. DOIPubMedGoogle Scholar
- Schmid J, Voss E, Soll DR. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–43.PubMedGoogle Scholar
- Scherer S, Stevens DA. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–9.PubMedGoogle Scholar
- Soll DR. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000;13:332–70. DOIPubMedGoogle Scholar
- Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco: W.H. Freeman; 1973.
- Blignaut E, Pujol C, Lockhart S, Joly S, Soll DR. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa. J Clin Microbiol. 2002;40:826–36. DOIPubMedGoogle Scholar
- Ghannoum MA, Filler SG, Ibrahim AS, Fu Y, Edwards JE Jr. Modulation of interactions of Candida albicans and endothelial cells by fluconazole and amphotericin B. Antimicrob Agents Chemother. 1992;36:2239–44.PubMedGoogle Scholar
- De Bernardis F, Lorenzini R, Verticchio R, Agatensi L, Cassone A. Isolation, acid proteinase secretion, and experimental pathogenicity of Candida parapsilosis from outpatients with vaginitis. J Clin Microbiol. 1989;27:2598–603.PubMedGoogle Scholar
- Leidich SD, Ibrahim AS, Fu Y, Koul A, Jessup C, Vitullo J, Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J Biol Chem. 1998;73:26078–86. DOIPubMedGoogle Scholar
- Mirbod F, Banno Y, Ghannoum MA, Ibrahim AS, Nakashima S, Kitajima Y, Purification and characterization of lysophospholipase-transacylase (h-LPTA) from a highly virulent strain of Candida albicans. Biochim Biophys Acta. 1995;1257:181–8.PubMedGoogle Scholar
- Lin D, Wu LC, Rinaldi MG, Lehmann PF. Three distinct genotypes within Candida parapsilosis from clinical sources. J Clin Microbiol. 1995;33:1815–21.PubMedGoogle Scholar
- Ghannoum MA, Radwan SS. Candida adherence to epithelial cells. Boca Raton (FL): CRC Press; 1990.
- Rex JH, Walsh TJ, Sobel JD, Filler SG, Pappas PG, Dismukes WE, Practice guidelines for the treatment of candidiasis. Clin Infect Dis. 2000;30:662–78. DOIPubMedGoogle Scholar
- Vrioni G, Matsiota-Bernard P. Molecular typing of Candida isolates from patients hospitalized in an intensive care unit. J Infect. 2001;42:50–6. DOIPubMedGoogle Scholar
- Shin JH, Shin DH, Song JW, Kee SJ, Suh SP, Ryang DW. Electrophoretic karyotype analysis of sequential Candida parapsilosis isolates from patients with persistant or recurrent fungemia. J Clin Microbiol. 2001;39:1258–63. DOIPubMedGoogle Scholar
- De Bernardis F, Mondello F, San Millan R, Ponton J, Cassone A. Biotyping and virulence properties of skin isolates of Candida parapsilosis. J Clin Microbiol. 1999;37:3481–6.PubMedGoogle Scholar
- Panagoda GJ, Ellepola AN, Samaranayake LP. Adhesion of Candida parapsilosis to epithelial and acrylic surfaces correlates with cell surface hydrophobicity. Mycoses. 2001;44:29–35. DOIPubMedGoogle Scholar
- Shin JH, Kee SJ, Shin MG, Kim SH, Shin DH, Lee SK, Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J Clin Microbiol. 2002;40:1244–8. DOIPubMedGoogle Scholar
- Ibrahim AS, Filler SG, Sanglard D, Edwards JE Jr, Hube B. Secreted aspartyl proteinases and interactions of Candida albicans with human endothelial cells. Infect Immun. 1998;66:3003–5.PubMedGoogle Scholar
- De Bernardis F, Chiani P, Ciccozzi M, Pellegrini G, Ceddia T, D’Offizzi G, Elevated aspartic proteinase secretion and experimental pathogenicity of Candida albicans isolates from oral cavities of subjects infected with human immunodeficiency virus. Infect Immun. 1996;64:466–71.PubMedGoogle Scholar
- De Bernardis F, Cassone A, Sturtevant J, Calderone R. Expression of Candida albicans SAP1 and SAP2 in experimental vaginitis. Infect Immun. 1995;63:1887–92.PubMedGoogle Scholar
- Kantarcioglu AS, Yucel A. Phospholipase and protease activities in clinical Candida isolates with reference to the sources of strains. Mycoses. 2002;45:160–5. DOIPubMedGoogle Scholar
- Branchini ML, Pfaller MA, Rhine-Chalberg J, Frempong T, Isenberg HD. Genotypic variation and slime production among blood and catheter isolates of Candida parapsilosis. J Clin Microbiol. 1994;32:452–6.PubMedGoogle Scholar
- Fernanado PH, Panagoda GJ, Samaranayake LP. The relationship between the acid and alkaline phosphatase activity and the adherence of clinical isolates of Candida parapsilosis to human buccal epithelial cells. APMIS. 1999;107:1034–42. DOIPubMedGoogle Scholar
- Barrett-Bee K, Hayes Y, Wilson RG, Ryley JF. A comparison of phospholipase activity, cellular adherence and pathogenicity of yeasts. J Gen Microbiol. 1985;131:1217–21.PubMedGoogle Scholar
- Ripeau JS, Fiorillo M, Aumont F, Belhumeur P, de Repentigny L. Evidence for differential expression of Candida albicans virulence genes during oral infection in intact and human immunodeficiency virus type 1-transgenic mice. J Infect Dis. 2002;185:1094–102. DOIPubMedGoogle Scholar
- Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. Differential expression of Candida albicans phospholipase B (PLB1) under various environmental and physiological conditions. Microbiology. 2002;149:1–7.PubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 10, Number 6—June 2004
| EID Search Options |
|---|
|
|
|
|
|
|




Please use the form below to submit correspondence to the authors or contact them at the following address:
*Mahmoud A. Ghannoum, Center for Medical Mycology, Dept. of Dermatology, University Hospitals of Cleveland, LKSD 5028, 11100 Euclid Ave., Cleveland, OH 44106, USA; fax: 216-844-1076
Top