Volume 18, Number 7—July 2012
Dispatch
Calicivirus from Novel Recovirus Genogroup in Human Diarrhea, Bangladesh
Figure 1
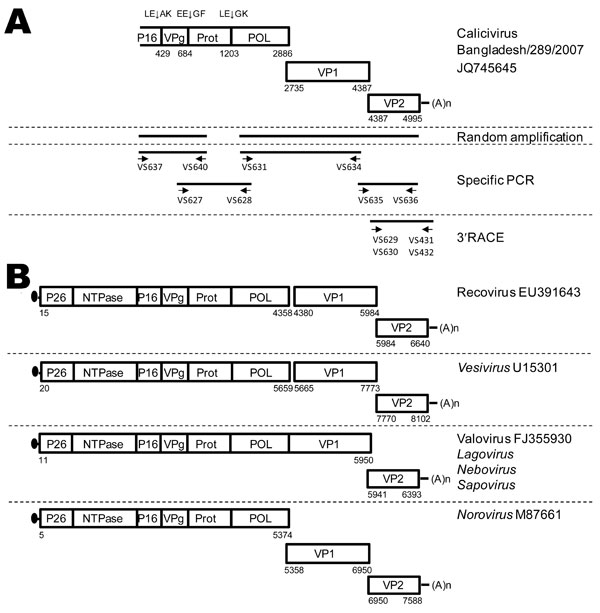
Figure 1. . Schematic outline of the strategies used for PCR amplification of calicivirus Bangladesh/289/2007. A) Schematic representation of the calicivirus Bangladesh/289/2007 genome. Boxes represent the open reading frames encoding the calicivirus proteins. Indicated are the poly(A)-tail (An); putative cleavage sites indicated by XX↓XX. The 5′ end of the genome was not obtained. The bottom of the panel shows a schematic outline of the reverse transcription PCRs employed to amplify calicivirus Bangladesh/289/2007 sequences by using random amplification, degenerate PCR, and 3′ rapid amplification of cDNA ends (RACE) PCR. The orientations and positions of the oligonucleotides on the calicivirus genome are depicted and sequences shown in Technical Appendix Table 2. B) Genome organization of caliciviruses in the genera Vesivirus, Nebovirus, Norovirus, Sapovirus, and Lagovirus and proposed genera Valovirus and Recovirus, for comparison with the new calicivirus Bangladesh/289/2007. The 5′ end of the genome is shown with a Vpg protein (black dots). Numbers indicate the nucleotide positions according to the virus genome for which the GenBank accession number is indicated.