Volume 21, Number 10—October 2015
Dispatch
Novel Paramyxoviruses in Bats from Sub-Saharan Africa, 2007–2012
Figure
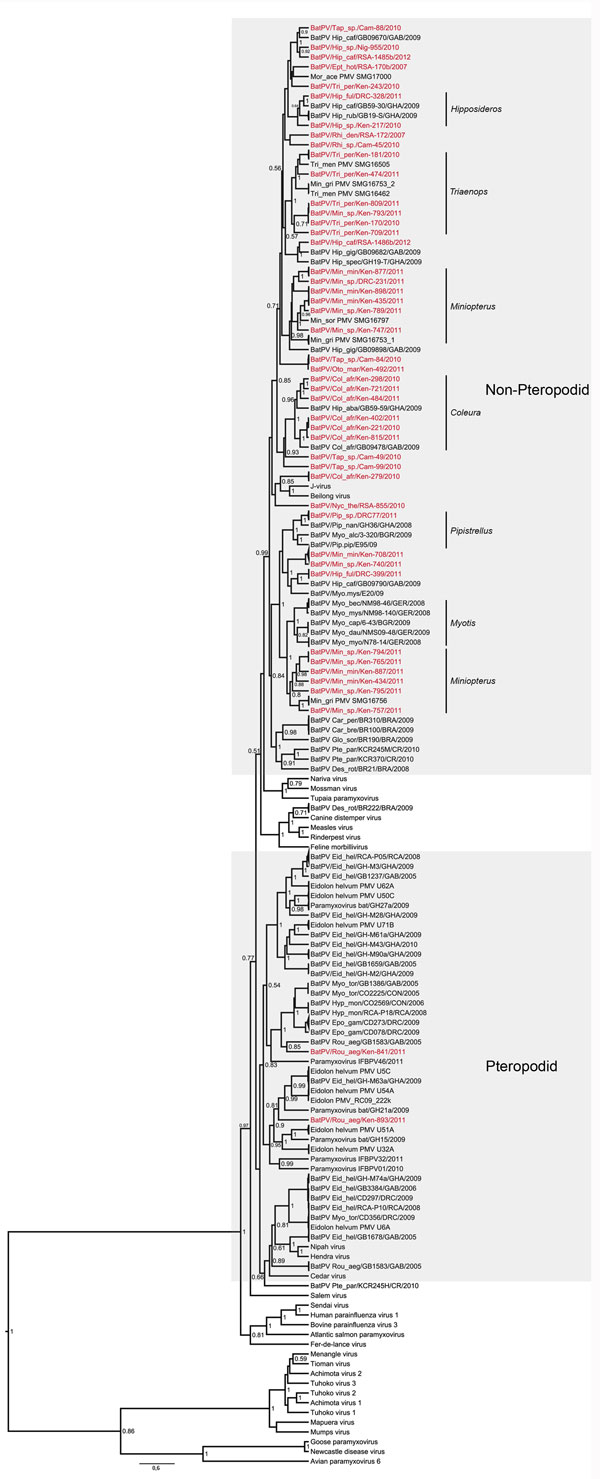
Figure. Maximum clade credibility tree based on partial polymerase (large) gene sequences (439 bp) of paramyxoviruses built in BEAST version 1.7.4 software (http://beast.bio.ed.ac.uk/), applying the general time reversible plus invariant sites plus gamma model inferred by jModelTest version 0.1.1 (10). Sequences detected in this study are indicated in red. Identical sequences were collapsed to only show a representative. Genus-specific clusters are indicated on the right and show possible opportunistic infections in other species grouping within these clusters. The 2 lineages reported are indicated in the gray areas.
References
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, Carstens EB, editors. Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press, Elsevier; 2011.
- Hooper P, Zaki S, Daniels P, Middleton D. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 2001;3:315–22. DOIPubMedGoogle Scholar
- Baker KS, Todd S, Marsh G, Fernandez-Loras A, Suu-Ire R, Wood JLN, Co-circulation of diverse paramyxoviruses in an urban African fruit bat population. J Gen Virol. 2012;93:850–6 and. DOIPubMedGoogle Scholar
- Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796.
- Wilkinson DA, Temmam S, Lebarbenchon C, Lagadec E, Chotte J, Guillebaud J, Identification of novel paramyxoviruses in insectivorous bats of the Southwest Indian ocean. Virus Res. 2012;170:159–63. DOIPubMedGoogle Scholar
- Drexler JF, Corman VM, Gloza-Rausch F, Seebens A, Annan A, Ipsen A, Henipavirus RNA in African bats. PLoS ONE. 2009;4:e6367. DOIPubMedGoogle Scholar
- Monadjem A, Taylor PJ, Cotterill FPD, Schoeman MC. Bats of southern and central Africa. Johannesburg (South Africa): Wits University Press; 2010.
- Tong S, Wang Chern S-W, Li W, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assay to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–8. DOIPubMedGoogle Scholar
- Weiss S, Nowak K, Fahr J, Wibbelt G, Mombouli J-V, Parra HJ, Henipavirus-related sequences in fruit bat bushmeat, Republic of Congo. Emerg Infect Dis. 2012;18:1536–7. DOIPubMedGoogle Scholar
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6. DOIPubMedGoogle Scholar
- Teeling EC, Springer MS, Madsen O, Bates P, O’Brien J, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–4. DOIPubMedGoogle Scholar
- Luby SP, Gurley ES, Hossain MJ. Transmission of human infection with Nipah virus. Clin Infect Dis. 2009;49:1743–8. DOIPubMedGoogle Scholar
- Wood JLN, Leach M, Waldman L, MacGregor H, Fooks AR, Jones KE, Framework for the study of zoonotic disease emergence and its drivers: spillover of bat pathogens as a case study. Philos Trans R Soc Lond B Biol Sci. 2012;367:2881–92. DOIPubMedGoogle Scholar
Page created: September 22, 2015
Page updated: September 22, 2015
Page reviewed: September 22, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.