Volume 22, Number 5—May 2016
Dispatch
Increased Rotavirus Prevalence in Diarrheal Outbreak Precipitated by Localized Flooding, Solomon Islands, 2014
Figure 2
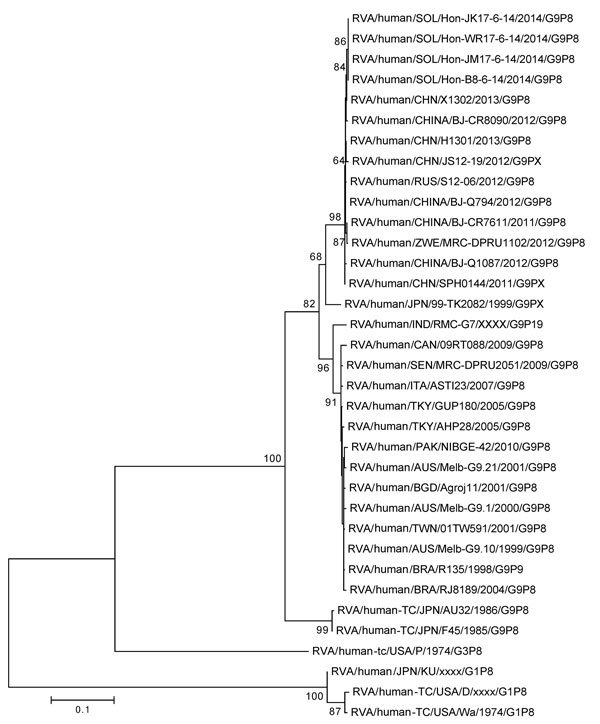
Figure 2. Nucleotide sequence–based phylogenetic tree of rotavirus viral protein (VP) 7 for isolates obtained in the Solomon Islands during an outbreak of diarrheal illness after flooding in the capital city of Honiara. Four isolates from different provinces had homologous VP7 sequences. We visually analyzed generated electropherograms and constructed contiguous DNA sequence files using the Sequencher Software program version 5.0.1 (Gene Codes Corp Inc., Ann Arbor, MI, USA). We performed nucleotide similarity searches using the BLAST (http://www.ncbi.nlm.nih.gov) and compared the nucleotide and deduced amino acid sequences of the VP7 gene with sequences available in GenBank possessing the entire open reading frame. We constructed multiple nucleotide and amino acid alignments using the MUSCLE algorithm in MEGA 6.0 (http://www.megasoftware.net/). Nucleotide and amino acid distance matrices were calculated by using the p-distance algorithm in MEGA 6.0. We selected the optimal evolutionary model based on the Akaike information criterion (corrected) ranking implemented in jModelTest (GitHub, Heidelberg, Germany) and generated maximum-likelihood phylogenetic trees using the nucleotide substitution model TrN+Gamma 4+I in MEGA 6.0, and assessed the robustness of branches by bootstrap analysis using 1,000 pseudoreplicate runs. Scale bar indicates base substitutions per site.