Volume 23, Number 6—June 2017
Dispatch
Seoul Virus Infection in Humans, France, 2014–2016
Figure 1
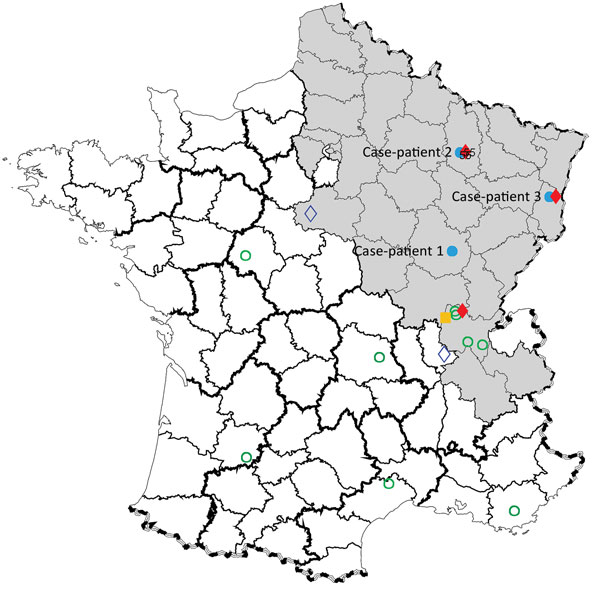
Figure 1. Geographic distribution of Seoul virus (SEOV) infections among human and rats, France 2016. Gray shading area indicates area of France to which Puumala virus is endemic. Open green circles indicate SEOV serologically confirmed human infections with SEOV reported by Ragnaud et al. (10), Le Guenno (11), and Bour et al. (6); solid blue circles indicate virologically confirmed human infections with SEOV reported in this study (patients 1, 2, and 3); solid yellow square indicates virologically confirmed human infection with SEOV reported by Macé al. (5); solid red diamonds indicate virologically confirmed SEOV infections in brown rats reported in this study; and open blue diamonds indicate virologically confirmed SEOV infections in brown rats reported by Heyman et al. (12) and Dupinay et al. (13).
References
- Clement J, Heyman P, McKenna P, Colson P, Avsic-Zupanc T. The hantaviruses of Europe: from the bedside to the bench. Emerg Infect Dis. 1997;3:205–11. DOIPubMedGoogle Scholar
- Kariwa H, Yoshimatsu K, Arikawa J. Hantavirus infection in East Asia. Comp Immunol Microbiol Infect Dis. 2007;30:341–56. DOIPubMedGoogle Scholar
- Knust B, Rollin PE. Twenty-year summary of surveillance for human hantavirus infections, United States. Emerg Infect Dis. 2013;19:1934–7. DOIPubMedGoogle Scholar
- Jameson LJ, Taori SK, Atkinson B, Levick P, Featherstone CA, van der Burgt G, et al. Pet rats as a source of hantavirus in England and Wales, 2013. Euro Surveill. 2013;18:20415.PubMedGoogle Scholar
- Macé G, Feyeux C, Mollard N, Chantegret C, Audia S, Rebibou JM, et al. Severe Seoul hantavirus infection in a pregnant woman, France, October 2012. Euro Surveill. 2013;18:20464.PubMedGoogle Scholar
- Bour A, Reynes JM, Plaisancie X, Dufour JF. [Seoul hantavirus infection-associated hemorrhagic fever with renal syndrome in France: A case report] [in French]. Rev Med Interne. 2016;37:493–6. DOIPubMedGoogle Scholar
- Klempa B, Fichet-Calvet E, Lecompte E, Auste B, Aniskin V, Meisel H, et al. Hantavirus in African wood mouse, Guinea. Emerg Infect Dis. 2006;12:838–40. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Centre National de Reference Hantavirus. Institut Pasteur. Annual activity report, 2015 [in French] [cited 2017 Feb 27]. https://www.pasteur.fr/fr/sante-publique/CNR/les-cnr/hantavirus/rapports-d-activite
- Ragnaud JM, Lamouliatte H, Paix MA, Fleury H, Roy J, Magnol R. [Hemorrhagic fever with renal syndrome: a case with jaundice] [in French]. Gastroenterol Clin Biol. 1986;10:686.PubMedGoogle Scholar
- Heyman P, Plyusnina A, Berny P, Cochez C, Artois M, Zizi M, et al. Seoul hantavirus in Europe: first demonstration of the virus genome in wild Rattus norvegicus captured in France. Eur J Clin Microbiol Infect Dis. 2004;23:711–7. DOIPubMedGoogle Scholar
- Dupinay T, Pounder KC, Ayral F, Laaberki MH, Marston DA, Lacôte S, et al. Detection and genetic characterization of Seoul virus from commensal brown rats in France. Virol J. 2014;11:32. DOIPubMedGoogle Scholar
- Lagerqvist N, Hagström Å, Lundahl M, Nilsson E, Juremalm M, Larsson I, et al. Molecular diagnosis of Puumala virus-caused hemorrhagic fever with renal syndrome. J Clin Microbiol. 2016;54:1335–9. DOIPubMedGoogle Scholar
- Mähler Convenor M, Berard M, Feinstein R, Gallagher A, Illgen-Wilcke B, Pritchett-Corning K, et al.; FELASA working group on revision of guidelines for health monitoring of rodents and rabbits. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab Anim. 2014;48:178–92. DOIPubMedGoogle Scholar