Volume 24, Number 1—January 2018
Research
Drug-Resistant Polymorphisms and Copy Numbers in Plasmodium falciparum, Mozambique, 2015
Abstract
One of the fundamental steps toward malaria control is the use of antimalarial drugs. The success of antimalarial treatment can be affected by the presence of drug-resistant populations of Plasmodium falciparum. To assess resistance, we used molecular methods to examine 351 P. falciparum isolates collected from 4 sentinel sites in Mozambique for K13, pfmdr1, pfcrt, and pfdhps polymorphisms and for plasmepsin2 (pfpm2) and pfmdr1 copy numbers. We found multiple copies of pfpm2 in 1.1% of isolates. All isolates carried K13 wild-type alleles (3D7-like), except 4 novel polymorphisms (Leu619Leu, Phe656Ile, Val666Val, Gly690Gly). Prevalence of isolates with pfcrt mutant (K76T) allele was low (2.3%). Prevalence of isolates with pfdhps mutant alleles (A437G and K540E) was >80%, indicating persistence of sulfadoxine/pyrimethamine resistance; however, markers of artemisinin were absent, and markers of piperaquine resistance were low. Piperaquine resistance isolates may spread in Mozambique as dihydroartemisinin/piperaquine drug pressure increases.
During the past decade, malaria control strategies have substantially reduced the malaria burden worldwide; several countries are advancing toward malaria elimination (1,2). A fundamental pillar for contributing to the reduction of the malaria burden has been artemisinin-based combination therapy. Unfortunately, the effectiveness of antimalarial drugs used for malaria treatment and chemoprevention during pregnancy has been threatened by the emergence of drug-resistant parasite populations (2–5).
The emergence of artemisinin resistance in Plasmodium falciparum, with reduced in vivo susceptibility to artesunate, was reported in Southeast Asia (3,6). Detectable polymorphisms in the Kelch 13 (K13) propeller domain in P. falciparum associated with artemisinin resistance have subsequently provided an additional tool for monitoring resistance to antimalarial drugs (7,8). In Cambodia, polymorphisms in the K13 propeller domain (mainly Y493H, R539T, I543T, and C580Y) were associated with in vitro prolonged parasite survival rates and in vivo delayed parasite clearance rates (8,9). Recently, plasmepsin 2 (pfpm2) copy number and pfcrt C101F polymorphism have been associated with piperaquine resistance (10–12). In addition, increased pfmdr1 copies have been associated with resistance to mefloquine (in vivo, in vitro, or both) and partially to lumefantrine (13–18). Specific point polymorphisms (at codons 86, 184, 1034, 1042, and 1246) of the pfmdr1 gene have also been linked to resistance to antimalarial drugs (19,20). In field isolates tested in vitro as well as in laboratory lines, N86Y polymorphism was associated with chloroquine resistance (21). Further, polymorphisms in the pfcrt gene have also been shown to affect parasite susceptibility to chloroquine (22), amodiaquine (23,24), and artemether/lumefantrine (25). Recently, a nonsynonymous polymorphism in the pfcrt gene was shown to be prevalent in the genetic background of K13 mutant artemisinin-resistant isolates (26). In addition, polymorphisms in pfdhfr and pfdhps genes, specifically the quintuple mutant, including the pfdhfr substitutions N51I, C59R, and S108N, as well as the pfdhps substitutions A437G and K540E, have been associated with a failure of sulfadoxine/pyrimethamine treatment against uncomplicated P. falciparum malaria (27). In Africa, the pfdhps K540E polymorphism has been considered a useful epidemiologic marker of the quintuple mutations (28).
The development of drug resistance could be influenced by multiple factors such as polymorphism rate, fitness costs, overall parasite load, strength of drug selection, treatment compliance, transmission intensity, host immunity, and erythrocyte disorders (29–31). Naturally acquired immunity plays a major role in the emergence and clearance of artemisinin-resistant parasites (32). Because of increasing concern over the effectiveness of the nationally recommended antimalarial drugs, the Mozambique Ministry of Health has made several changes in antimalarial drug policy. In 2002, chloroquine monotherapy was replaced with sulfadoxine/pyrimethamine/amodiaquine as the first line of treatment against uncomplicated malaria (33); 2 years later, this combination was replaced with artesunate/sulfadoxine/pyrimethamine (33). In 2008, artemether/lumefantrine was introduced to replace artesunate/sulfadoxine/pyrimethamine (34). Molecular markers for antimalarial drug resistance have been considered useful for confirming parasite resistance, a major factor causing treatment failure. To determine whether parasites carrying these polymorphisms or gene amplifications exist in Mozambique, we conducted molecular surveillance targeting K13, pfmdr1, pfcrt, and pfdhps polymorphisms and pfpm2 and pfmdr1 copy numbers in field isolates collected from 4 sentinel sites.
Study Sites and Population
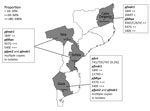
We performed a descriptive observational study on blood samples collected before artemether/lumefantrine treatment (on day 0) in 2015 from 352 symptomatic children at 4 sentinel sites in Mozambique (Figure 1): 1) Hospital Rural de Montepuez in Cabo Delgado Province (northern region), 2) Centro de Saúde de Dondo in Sofala Province (central region), 3) Hospital Provincial de Moatize in Tete Province (central region), and 4) Hospital Rural de Chokwe in Gaza Province (southern region). In Mozambique, transmission usually peaks during the rainy season (November–April). Transmission intensity in southern Mozambique is generally low, although areas of high transmission may still occur (35). To determine molecular markers of drug resistance, we analyzed samples collected during a clinical trial conducted in 2015 (registration no. ACTRN12616001680459); the trial aimed to assess the efficacy and safety of artemether/lumefantrine for treatment of uncomplicated P. falciparum malaria in children <5 years of age. The National Mozambican Ethical Review Committee (Mozambique) and Hospital Clínic (Barcelona, Spain) ethics review committees approved the study, and signed written informed consent was obtained from all participants’ guardian or parent.
Molecular Procedures
We extracted DNA from half of a 50-μL dried blood drop on Whatman 3-mm filter paper by using a QIAamp DNA Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. We used an ABI PRISM 7500 HT Real-Time System (Applied Biosystems, Foster City, CA, USA) to amplify purified parasite DNA templates, following a previously described method (36,37). A standard curve was prepared from an in vitro culture of 3D7 strain containing known numbers of ring-infected erythrocytes. The standard curve was run in triplicate for each test with 5 serially diluted points. Parasitemia in the clinical samples was quantified by extrapolation against the standard curve.
To assess polymorphisms in the K13, pfcrt, pfmdr1, and pfdhps genes, we amplified purified DNA templates by using a 2720 Thermal Cycler (Applied Biosystems), following protocols described for K13 PCR (38) and pfcrt PCR (35). To genotype polymorphisms in pfmdr1 and pfdhps genes, we designed new assays by using Sanger sequencing and restriction fragment length polymorphisms (Technical Appendix). A total of 6 positive controls with known K13 alleles, provided by the Institut Pasteur in Cambodia, and 4 parasite lines (3D7, 7G8, Dd2, and V1/S) with known pfcrt and pfmdr1 alleles, available in the laboratory, were also processed, amplified, and sequenced at the same time as the studied samples (PCR characteristics in Technical Appendix Table). To determine the detection limit of Sanger sequencing, we used artificially mixed DNA samples of P. falciparum laboratory strains containing various known proportions of wild- and mutant-type alleles of pfcrt (K76T) and pfmdr1 (Y184F and S1034C) genes. To estimate polymorphism frequency, we considered isolates with mixed alleles to be mutated.
We assessed copy numbers of pfpm2 and pfmdr1 genes as described elsewhere (11) with minor changes (Technical Appendix) by using quantitative PCR (qPCR). We performed amplification in 20-μL reaction mixtures for pfpm2, pfmdr1, and pfβ-tubulin genes, separately. We used the pfβ-tubulin gene as an endogenous control. All samples with estimated copy numbers >1.5 were defined as containing multiple copies and repeated for confirmation. The estimated copy numbers were the average of the copy number of each clone in the isolate.
Data Analyses
We calculated the proportion of the mutant alleles and isolates with multiple copies of pfpm2 and pfmdr1 genes on the basis of the number of samples with wild- and mutant-type alleles as well as isolates with single and multiple copies of the gene from P. falciparum isolates from each study site. To compare continuous data and categorical data between sites, respectively, we performed analyses of variance and χ2 tests. We defined statistical significance as p<0.05.
Demographics and P. falciparum Infection
Among the 352 blood samples collected before artemether/lumefantrine treatment (on day 0) during 2015, and followed up as part of the clinical trial, 351 (99.7%) were P. falciparum–infection positive according to 18SrRNA qPCR. The mean (± SD) parasitemia (by qPCR) was 100,229 ± 325,214 parasites/μL. Among participants, 159 (45.2%) were female, mean (± SD) age was 2.8 ± 1.3 y, mean body temperature was 38.1 ± 1.1°C, and mean hemoglobin level was 9.2 ± 1.9 g/dL. We also compared demographic data and parasite densities according to study site (Table 1). Efficacy of artemether/lumefantrine in the in vivo study was high, and for nearly all patients (349 [99.4%] of 351), parasitemia reverted to 0 in the first 3 days; however, for 2 patients, parasites were still detectable by microscopy: 1 from Moatize (514 parasites/μL) and 1 from Chokwe (3,763 parasites/μL). PCRs targeting msp1, msp2, and glurp genes were used to differentiate recrudescence (same parasite strain) and reinfection (different parasite strain). We noted recrudescence of P. falciparum infections for 5 children (1 in Chokwe, 3 in Moatize, and 1 in Montepuez) on days 21 and 28 after artemether/lumefantrine administration and reinfection for 7 children (3 in Moatize and 4 in Montepuez); 3 were reinfected on day 21 and 4 on day 28 (39).
The polymorphism analyses of K13, pfmdr1, pfcrt, and pfdhps genes were successful for 98.3% to 100% isolates. Because no amplifications were noticed in negative controls (with water and human genomic DNA), PCR assays were specific to P. falciparum genomic DNA only.
Detection Limit of Mixed Samples by Sanger Sequencing
We identified ¨A¨ alleles of pfcrt (K76T) and pfmdr1 (Y184F) codons in artificially mixed samples by using Sanger sequencing when the proportion of target DNA was >10%. However, we identified ¨C¨ and ¨T¨ alleles of pfcrt (K76T) and pfmdr1 (Y184F) polymorphisms, respectively, in mixed samples when their proportion was >20% (Technical Appendix Figure 1). For pfmdr1 (S1034C) polymorphism, the minor allele was detected when its proportion was >20% in a mixed sample (Technical Appendix Figure 1, panel C). For positive controls, we used several parasite lines with known K13, pfmdr1, and pfcrt alleles. As expected, sequencing analysis of all positive controls revealed wild- and mutant-type alleles of K13, pfmdr1, and pfcrt polymorphisms.
Copy Numbers for pfpm2 and pfmdr1
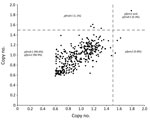
We successfully analyzed 351 (100%) samples for copy number variation in the pfpm2 and pfmdr1 genes. PCR efficiencies were 98.4% for pfpm2, 97.2% for pfmdr1, and 99.2% for pfβ-tubulin genes. As expected, the estimated pfpm2 and pfmdr1 copy numbers for the positive controls were 3–4 copies. The estimated mean (interquartile range) copy numbers were 3.51 (3.37–3.62) for pfpm2 and 3.62 (3.51–3.79) for pfmdr1 positive controls. When we used a copy number threshold of 1.5 to define multiple copies, only 4 (1.1%) and 5 (1.4%) of the 351 isolates had multiple copies of pfpm2 and pfmdr1, respectively (Table 2; Figure 2). The range of estimated pfpm2 copy numbers was 0.59–1.79 and of pfmdr1 was 0.58–1.88. The copy number of pfpm2 and pfmdr1 genes did not significantly differ between isolates from different sites. The proportion of isolates with multiple copies of the pfpm2 gene was the highest at Chokwe (2 [2.3%] of 87). Only 1 (1.1%) of 88 samples from Dondo had multiple copies of pfpm2 and pfmdr1 genes.
K13 Polymorphisms
We successfully achieved K13 PCR and sequencing for all 351 isolates. None of the isolates analyzed contained the polymorphisms most frequently found in isolates from Cambodia (8). However, we observed 4 novel polymorphisms at nt 1725147 (codon 619; 0.28% [1/351]); 1725032 (codon 656; 0.28% [1/351]); 1725000 (codon 666; 0.57% [2/351]); and 1724927 (codon 690; 0.85% [3/351]) of the K13 gene. All polymorphisms were synonymous except for 1 at codon 656, which led to a change from phenylalanine to isoleucine. When we compared frequencies of new polymorphisms between sites, we found no significant differences. We also observed the polymorphism Cys469Cys, previously described in P. falciparum field isolates from Ghana (40), in 3 (0.85%) of the 351 isolates. Isolates from patients with parasitemia on day 3 and recrudescence contained wild-type K13 gene polymorphisms.
pfcrt Polymorphisms
We successfully amplified all 351 samples for pfcrt and sequenced the amplification products; mutant alleles were found at codons M74I, N75E, and K76T only in 8 (2.3%) samples. The mutant alleles (M74I, N75E, and K76T) were present only in isolates collected from Chokwe (8 [9.2%] of 87). When we compared frequencies of mutant alleles between sites, the difference was significant (p<0.0001). In the studied isolates, the mutant (F) allele at codon 101 was absent. Isolates from patients with parasitemia on day 3 and recrudescence contained wild-type pfcrt gene polymorphisms.
pfmdr1 Polymorphisms
We successfully amplified and sequenced pfmdr1_f1 for 351 (100%) samples and pfmdr1_f2 fragments for 350 (99.7%) samples. We identified 15 polymorphisms all across the pfmdr1 gene, including 5 (33.3%) with nonsynonymous polymorphisms and 10 (66.7%) with synonymous polymorphisms. Among nonsynonymous polymorphisms, 3 (T1192A, F1194S, and Y1197N) were newly identified and 2 (N86Y and Y184F) had been previously reported (41). Among synonymous polymorphisms, 7 (L1030L, D1061D, D1127D, S1137S, L1174L, D1179D, and N1189N) were newly identified and 3 (G102G, G182G, and T1069T) had been previously reported (41). Among the 351 isolates, we found 11 (3.1%) N86Y and 164 (46.7%) Y184F mutant alleles (Table 3). All newly identified nonsynonymous mutant alleles were present only once, except for Y1197N, which was found twice (0.6% [2/350]). The frequency of polymorphisms (N86Y and D1179D) differed significantly between isolates from the 4 sites (Table 3). The proportion of N86Y and D1179D polymorphisms was highest in isolates from Chokwe. We observed none of the other most frequent polymorphisms (S1034C, N1042D, and D1246Y) of the pfmdr1 gene among the analyzed samples. Isolates from patients with parasitemia on day 3 and recrudescence contained wild-type pfmdr1 gene polymorphisms.
pfdhps Polymorphisms
Polymorphism analysis by PCR followed by sequencing for S436F and A437G polymorphisms was successful for 345 (98.3%) samples and analysis by PCR–restricted fragment length polymorphism for K540E polymorphism for 348 (99.1%) samples. Among all isolates, 10 (2.9%) of 345 contained S436F, 289 (83.8%) of 345 contained A437G, and 286 (82.2%) of 348 contained K540E mutant alleles. At codon 436, we also found 3 mutant alleles: S436C (0.9%), S436A (4.9%), and S436H (0.6%). When we compared frequencies of 3 single-nucleotide polymorphisms at different sites, we noted significant differences (Table 4). The proportion of isolates with A437G and K540E polymorphisms was the highest at Moatize, and the proportion with S436F, S436C, S436A, and S436H alleles was highest at Montepuez.
We provide evidence for the presence of multiple copies of pfpm2 in 4 (1.1%) of 351 P. falciparum isolates circulating in southern Mozambique despite the absence of piperaquine drug pressure. Thus, with adequate drug pressure, isolates resistant to piperaquine may spread in Mozambique, as occurred in Southeast Asia (10,42,43). In selected areas of Cambodia in 2008, piperaquine was introduced as a partner drug of artemisinin (44). Soon after its introduction, as early as 2010, piperaquine resistance in western Cambodia emerged at an alarming rate (45). Subsequent reports confirmed a rapid increase in failure of dihydroartemisinin/piperaquine in other parts of Cambodia (42,46,47). The most frequent K13 mutants associated with artemisinin resistance were absent in the isolates from Mozambique. We also determined that prevalence of pfcrt (K76T) and pfmdr1 (N86Y) markers of resistance are low, supporting previous evidence for the return of parasites carrying pfcrt wild-type alleles in Mozambique (35), in contrast to persistence of pfdhps (A437G [83.8%]) and K540E [82.2%]) polymorphisms, markers of sulfadoxine/pyrimethamine resistance (34). The well-characterized polymorphism in pfmdr1 (Y184F [46.7%]) was also prevalent in Mozambique.
We found very low prevalence (<1%) for 4 new polymorphisms (Leu619Leu, Phe656Ile, Val666Val, and Gly690Gly) in the K13 gene of P. falciparum isolates from Mozambique. All polymorphisms except Phe656Ile were synonymous. Previously, V494I K13 nonsynonymous polymorphism has also been reported in Mozambique (48). In Africa, K13 nonsynonymous polymorphisms have also been reported at low frequencies in isolates from Cameroon, Central African Republic, Democratic Republic of the Congo, Gabon, The Gambia, Kenya, Madagascar, Malawi, Mali, Rwanda, Togo, Uganda, Zambia, and Equatorial Guinea (38,40,49–50; references 51,52 in Technical Appendix). The association of nonsynonymous polymorphisms with delayed parasite clearance has only recently been identified in Africa (reference 52 in Technical Appendix).
Resistance to both chloroquine and amodiaquine has been mainly associated with a single K76T mutant allele in the pfcrt gene (22–24). In our study, its prevalence in 8 (2.3%) of 351 samples was significantly lower than that found in previous studies in Mozambique (33,34; reference 53 in online Technical Appendix). Our pfcrt data align with previous evidence for the return of parasites carrying pfcrt wild-type alleles in Mozambique (35) and in other countries in Africa, such as Ethiopia (reference 54 in Technical Appendix), Malawi (reference 55 in Technical Appendix), and Cameroon (reference 56 in Technical Appendix). The selective disadvantage of mutant parasites in the absence of drug pressure has been proposed as the leading factor contributing to the reemergence of chloroquine-susceptible parasites (reference 57 in Technical Appendix). Because artemether/lumefantrine has been shown to select for the wild-type pfcrt 76K allele (25), this reemergence might be accelerated because of the increased use of artemether/lumefantrine as a first-line treatment for uncomplicated malaria in Mozambique (reference 53 in Technical Appendix).
Our study also provides evidence for the presence of few P. falciparum isolates with multiple copies of the pfmdr1 gene (5 [1.4%] of 351) circulating in southern Mozambique (34). Increased pfmdr1 copies have been associated with resistance to mefloquine and partial resistance to lumefantrine (13–18). Our study found that prevalence of the pfmdr1 N86Y mutant allele has decreased and the Y184F mutant allele has increased over time, in contrast with findings of other studies from Mozambique (34; references 55,58 in Technical Appendix). We identified 10 new polymorphisms (L1030L, D1061D, D1127D, S1137S, L1174L, D1179D, N1189N, T1192A, F1194S, and Y1197N) that had not been previously described for the pfmdr1 gene. Among the 15 polymorphisms identified in the pfmdr1 gene, we observed significant differences between sites for the N86Y and D1179D polymorphisms only.
Of 351 children who had received adequate treatment with artemether/lumefantrine (6 doses), 2 were still positive for parasitemia on day 3 (39). These isolates contained wild-type K13 gene polymorphisms. P. falciparum–positive patients for whom artemether/lumefantrine treatment failed had parasites that carried wild-type pfcrt and pfmdr1 polymorphisms. This observation suggests that in vivo artemether/lumefantrine resistance may be caused not only by variations in the pfcrt and pfmdr1 genes but possibly by parasite selection of variations in other genes; however, drug bioavailability issues may also have contributed.
A high proportion of the P. falciparum isolates from Mozambique contained K540E (82.2%) and A437G (83.8%) mutant alleles. These mutant alleles may still not jeopardize the effectiveness of sulfadoxine/pyrimethamine for malaria prevention in Mozambique; recent findings suggest that only >90% prevalence of a pfdhps K540E polymorphism could reduce the effectiveness of intermittent preventive therapy to clear peripheral parasites and prevent new infections during pregnancy (reference 59 in Technical Appendix). Therefore, sulfadoxine/pyrimethamine remains effective for intermittent preventive therapy during pregnancy, despite the high frequency of quintuple mutants; thus, the World Health Organization continues to recommend the use of intermittent preventive therapy to prevent malaria during pregnancy (references 60–62 in Technical Appendix). However, alternative antimalarial drugs for intermittent preventive therapy during pregnancy are needed because the prevalence of the K540E polymorphism in Mozambique is close to the threshold.
In conclusion, we report that prevalence of isolates with multiple copies of pfpm2 is lower than that found by previous studies in Cambodia (34.3%) and Vietnam (54.3%) (10,43), and we report the absence of K13 polymorphisms known to be associated with artemisinin resistance. We also report the return of parasites carrying pfcrt wild-type alleles (except in Chokwe) and persistence of parasites with pfdhps mutations associated with sulfadoxine/pyrimethamine resistance in Mozambique. Sulfadoxine/pyrimethamine–resistant isolates may be maintained by the constant use of intermittent preventive therapy during pregnancy, use of drug outside of hospitals, the very common use of co-trimoxazole (as prophylaxis for HIV-infected persons), and the low fitness cost of the polymorphisms (33; references 63,64 in Technical Appendix). In contrast, the fitness cost of the pfcrt mutant allele seems to be high, probably accounting for the return of parasites carrying pfcrt wild-type alleles in Mozambique (reference 57 in Technical Appendix). Current regional elimination efforts, as part of the G8 Malaria Elimination Initiative, may lead to more aggressive strategies involving population-wide distribution of antimalarial drugs, such as dihydroartemisinin/piperaquine, resulting in significantly increased drug pressure. Our findings might provide baseline prevalence data that enable us to directly determine the effects that increasing malaria control efforts or elimination programs will have on resistance evolution.
Dr. Gupta is a postdoctoral fellow at the ISGlobal, Barcelona Centre for International Health Research, Hospital Clínic, Universitat de Barcelona, Barcelona, Spain. His research focus is use of molecular tools for the active surveillance of emerging drug resistance, gene deletions, and afebrile malaria in malaria-endemic regions.
Acknowledgments
We thank all study participants, field workers, laboratory workers, and everyone who supported this study directly or indirectly. We also thank Silvie Huijben, Mercedes Rubio, and Nilo Ortiz de Zugasti Carrón for their useful comments on this manuscript.
We thank the World Health Organization for providing the funds for this study, as well as the Instituto de Salud Carlos III (PI13/01478 cofunded by the Fondo Europeo de Desarrollo Regional, and CES10/021-I3SNS) to A.M. A.M. is also supported by the Departament d’Universitats i Recerca de la Generalitat de Catalunya, Agència de Gestió d'Ajuts Universitaris i de Recerca (2014SGR263). H.G. has a fellowship from the Overseas Postdoctoral Fellowship program by the Science and Engineering Research Board, Department of Science & Technology, Government of India (SB/OS/PDF-043/2015-16). The Centro de Investigaçao em Saude de Manhica receives major core funding from the Spanish Agency for International Cooperation. ISGlobal is a member of the CERCA Programme, Generalitat de Catalunya.
M.W. and P.R. are staff members of the World Health Organization. These authors alone are responsible for the views expressed in this publication, and they do not necessarily represent the decisions, policy, or views of the World Health Organization.
References
- O’Meara WP, Mangeni JN, Steketee R, Greenwood B. Changes in the burden of malaria in sub-Saharan Africa. Lancet Infect Dis. 2010;10:545–55. DOIPubMedGoogle Scholar
- World Health Organization. World malaria report 2016 [cited 2017 Mar 24]. http://apps.who.int/iris/bitstream/10665/252038/1/9789241511711-eng.pdf?ua=1
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67. DOIPubMedGoogle Scholar
- Harrington WE, Mutabingwa TK, Kabyemela E, Fried M, Duffy PE. Intermittent treatment to prevent pregnancy malaria does not confer benefit in an area of widespread drug resistance. Clin Infect Dis. 2011;53:224–30. DOIPubMedGoogle Scholar
- Hamer DH, Ndhlovu M, Zurovac D, Fox M, Yeboah-Antwi K, Chanda P, et al. Improved diagnostic testing and malaria treatment practices in Zambia. JAMA. 2007;297:2227–31. DOIPubMedGoogle Scholar
- Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM; Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20. DOIPubMedGoogle Scholar
- World Health Organization. Artemisinin and artemisinin-based combination therapy resistance. Status report. 2017 [cited 2017 May 5]. http://apps.who.int/iris/bitstream/10665/255213/1/WHO-HTM-GMP-2017.9-eng.pdf?ua=1
- Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5. DOIPubMedGoogle Scholar
- Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al.; Tracking Resistance to Artemisinin Collaboration (TRAC). Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–23. DOIPubMedGoogle Scholar
- Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis. 2017;17:174–83. DOIPubMedGoogle Scholar
- Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis. 2017;17:164–73. DOIPubMedGoogle Scholar
- Dhingra SK, Redhi D, Combrinck JM, Yeo T, Okombo J, Henrich PP, et al. A variant PfCRT isoform can contribute to Plasmodium falciparum resistance to the first-line partner drug piperaquine. MBio. 2017;8:e00303–17. DOIPubMedGoogle Scholar
- Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–35. DOIPubMedGoogle Scholar
- Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. DOIPubMedGoogle Scholar
- Lim P, Alker AP, Khim N, Shah NK, Incardona S, Doung S, et al. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11. DOIPubMedGoogle Scholar
- Price RN, Cassar C, Brockman A, Duraisingh M, van Vugt M, White NJ, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–9.PubMedGoogle Scholar
- Wilson CM, Volkman SK, Thaithong S, Martin RK, Kyle DE, Milhous WK, et al. Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol. 1993;57:151–60. DOIPubMedGoogle Scholar
- Gil JP, Krishna S. pfmdr1 (Plasmodium falciparum multidrug drug resistance gene 1): a pivotal factor in malaria resistance to artemisinin combination therapies. Expert Rev Anti Infect Ther. 2017;15:527–43. DOIPubMedGoogle Scholar
- Kamugisha E, Jing S, Minde M, Kataraihya J, Kongola G, Kironde F, et al. Efficacy of artemether-lumefantrine in treatment of malaria among under-fives and prevalence of drug resistance markers in Igombe-Mwanza, north-western Tanzania. Malar J. 2012;11:58. DOIPubMedGoogle Scholar
- Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–5. DOIPubMedGoogle Scholar
- Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–90. DOIPubMedGoogle Scholar
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–71. DOIPubMedGoogle Scholar
- Folarin OA, Bustamante C, Gbotosho GO, Sowunmi A, Zalis MG, Oduola AM, et al. In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop. 2011;120:224–30. DOIPubMedGoogle Scholar
- Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A. 2009;106:18883–9. DOIPubMedGoogle Scholar
- Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199:750–7. DOIPubMedGoogle Scholar
- Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–34. DOIPubMedGoogle Scholar
- Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8. DOIPubMedGoogle Scholar
- World Health Organization. Policy recommendation on intermittent preventive treatment during infancy with sulfadoxine-pyrimethamine (SP-IPTi) for Plasmodium falciparum malaria control in Africa. Geneva: The Organization. p. 1–3.
- Petersen I, Eastman R, Lanzer M. Drug-resistant malaria: molecular mechanisms and implications for public health. FEBS Lett. 2011;585:1551–62. DOIPubMedGoogle Scholar
- Hastings IM, Watkins WM. Intensity of malaria transmission and the evolution of drug resistance. Acta Trop. 2005;94:218–29. DOIPubMedGoogle Scholar
- Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–92. DOIPubMedGoogle Scholar
- Ataide R, Ashley EA, Powell R, Chan JA, Malloy MJ, O’Flaherty K, et al. Host immunity to Plasmodium falciparum and the assessment of emerging artemisinin resistance in a multinational cohort. Proc Natl Acad Sci U S A. 2017;114:3515–20. DOIPubMedGoogle Scholar
- Enosse S, Magnussen P, Abacassamo F, Gómez-Olivé X, Rønn AM, Thompson R, et al. Rapid increase of Plasmodium falciparum dhfr/dhps resistant haplotypes, after the adoption of sulphadoxine-pyrimethamine as first line treatment in 2002, in southern Mozambique. Malar J. 2008;7:115. DOIPubMedGoogle Scholar
- Raman J, Mauff K, Muianga P, Mussa A, Maharaj R, Barnes KI. Five years of antimalarial resistance marker surveillance in Gaza Province, Mozambique, following artemisinin-based combination therapy roll out. PLoS One. 2011;6:e25992. DOIPubMedGoogle Scholar
- Galatas B, Nhamussua L, Candrinho B, Mabote L, Cisteró P, Gupta H, et al. In-vivo efficacy of chloroquine to clear asymptomatic infections in Mozambican adults: a randomized, placebo-controlled trial with implications for elimination strategies. Sci Rep. 2017;7:1356. DOIPubMedGoogle Scholar
- Mayor A, Serra-Casas E, Bardají A, Sanz S, Puyol L, Cisteró P, et al. Sub-microscopic infections and long-term recrudescence of Plasmodium falciparum in Mozambican pregnant women. Malar J. 2009;8:9. DOIPubMedGoogle Scholar
- Taylor SM, Mayor A, Mombo-Ngoma G, Kenguele HM, Ouédraogo S, Ndam NT, et al. A quality control program within a clinical trial Consortium for PCR protocols to detect Plasmodium species. J Clin Microbiol. 2014;52:2144–9. DOIPubMedGoogle Scholar
- Ménard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, et al.; KARMA Consortium. KARMA Consortium. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–64. DOIPubMedGoogle Scholar
- Salvador C, Rafael B, Matsinhe F, Candrinho B, Muthemba R, De Carvalho E, et al. Efficacy and safety of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria at sentinel sites in Mozambique, 2015. Acta Trop. 2017;171:146–50. DOIPubMedGoogle Scholar
- Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2015;211:1352–5.PubMedGoogle Scholar
- Andriantsoanirina V, Ratsimbasoa A, Bouchier C, Jahevitra M, Rabearimanana S, Radrianjafy R, et al. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr-1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob Agents Chemother. 2009;53:4588–97. DOIPubMedGoogle Scholar
- Leang R, Taylor WR, Bouth DM, Song L, Tarning J, Char MC, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59:4719–26. DOIPubMedGoogle Scholar
- Phuc BQ, Rasmussen C, Duong TT, Dong LT, Loi MA, Ménard D, et al. Treatment failure of dihydroartemisinin/piperaquine for Plasmodium falciparum malaria, Vietnam. Emerg Infect Dis. 2017;23:715–7. DOIPubMedGoogle Scholar
- Denis MB, Tsuyuoka R, Poravuth Y, Narann TS, Seila S, Lim C, et al. Surveillance of the efficacy of artesunate and mefloquine combination for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2006;11:1360–6. DOIPubMedGoogle Scholar
- Leang R, Barrette A, Bouth DM, Menard D, Abdur R, Duong S, et al. Efficacy of dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax in Cambodia, 2008 to 2010. Antimicrob Agents Chemother. 2013;57:818–26. DOIPubMedGoogle Scholar
- Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–65. DOIPubMedGoogle Scholar
- Chaorattanakawee S, Lon C, Jongsakul K, Gawee J, Sok S, Sundrakes S, et al. Ex vivo piperaquine resistance developed rapidly in Plasmodium falciparum isolates in northern Cambodia compared to Thailand. Malar J. 2016;15:519. DOIPubMedGoogle Scholar
- Escobar C, Pateira S, Lobo E, Lobo L, Teodosio R, Dias F, et al. Polymorphisms in Plasmodium falciparum K13-propeller in Angola and Mozambique after the introduction of the ACTs. PLoS One. 2015;10:e0119215. DOIPubMedGoogle Scholar
- World Health Organization. Status report on artemisinin and ACT resistance. 2015 [cited 2017 Mar 20]. http://www.who.int/malaria/publications/atoz/status-rep-artemisinin-resistance-sept2015.pdf
- Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH, et al. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis. 2015;21:490–2. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 24, Number 1—January 2018
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Alfredo Mayor, ISGlobal, Barcelona Ctr. Int. Health Res. (CRESIB), Hospital Clínic, Universitat de Barcelona, Carrer Rosselló 153 (CEK Bldg), E-08036 Barcelona, Spain
Top