Volume 24, Number 5—May 2018
Dispatch
Human Usutu Virus Infection with Atypical Neurologic Presentation, Montpellier, France, 2016
Figure 1
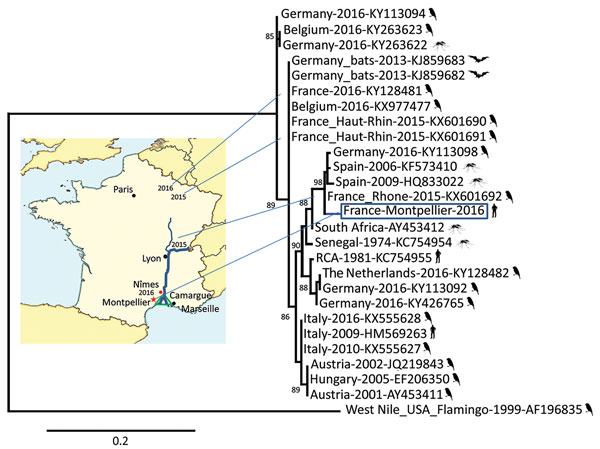
Figure 1. Phylogenetic relationship of the France-Montpellier-2016 strain of Usutu virus (USUV) (box; GenBank accession no. LT854220), isolated from a 39-year-old man in Montpellier, France, who had an atypical neurologic presentation, compared with other USUV strains based on the partial nonstructural protein 5 gene sequence. USUV sequences are shown with their country of isolation, year of isolation, and GenBank accession numbers. Hosts from which the strains were detected (bird, mosquito, bat, or human) are shown next to strain names. Analysis was processed through the French phylogeny website (http://www.phylogeny.fr). Nucleotide sequences were aligned by using MUSCLE software (https://www.ebi.ac.uk/Tools/msa/muscle/). The phylogenetic tree was constructed by using the maximum-likelihood method in PhyML (http://www.atgc-montpellier.fr/phyml/). One hundred bootstrap datasets with random sequence addition were computed to generate a consensus tree drawn with TreeDyn software (http://www.treedyn.org/) and rooted with a West Nile virus sequence (GenBank accession no. AF196835). Numbers along branches are bootstrap values. Map shows locations where USUV strains were detected in France during 2015–2016. Scale bar indicates nucleotide substitutions per site.