Volume 25, Number 10—October 2019
Research Letter
Lassa Virus in Pygmy Mice, Benin, 2016–2017
Figure
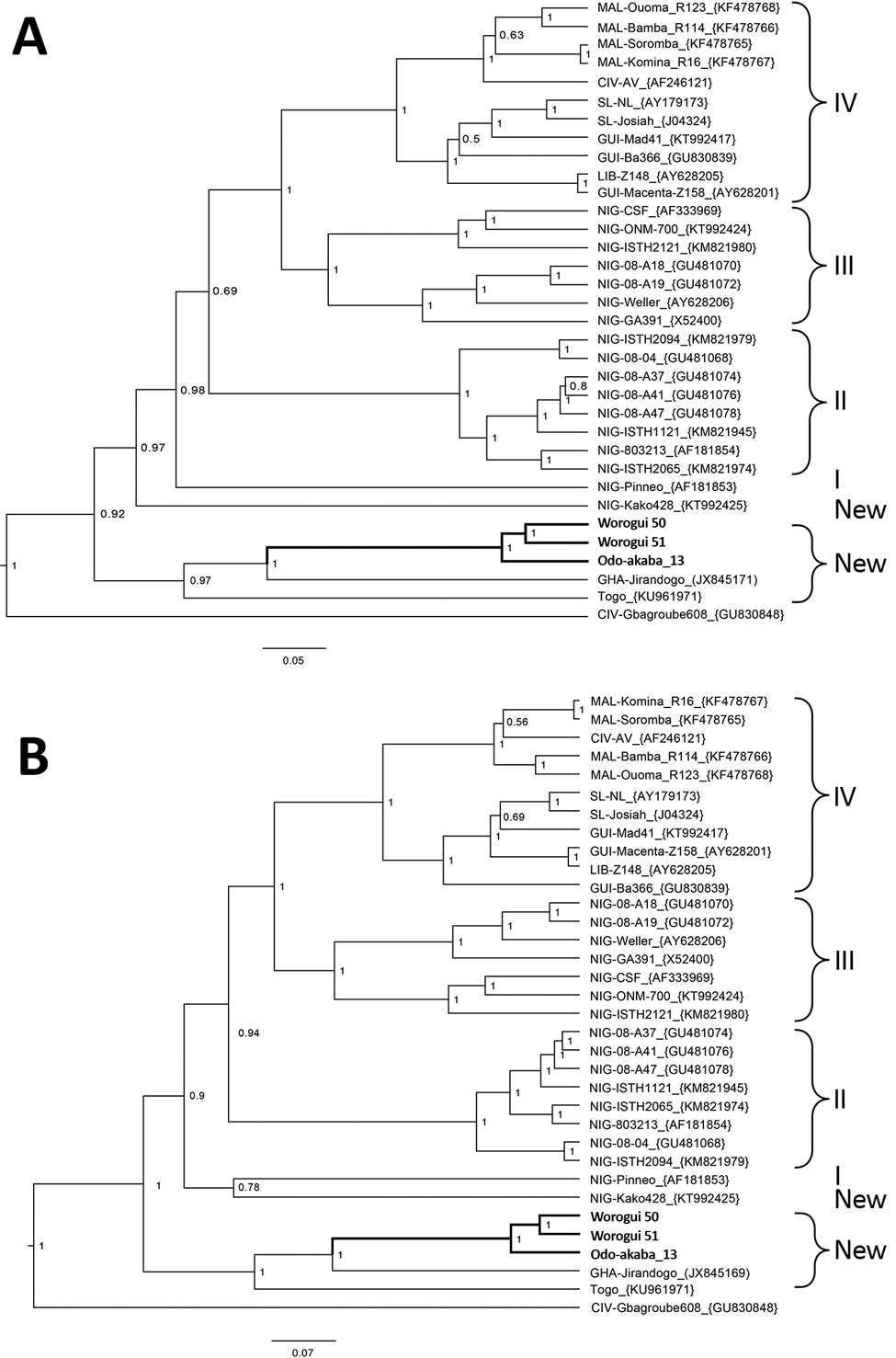
Figure. Bayesian phylogenetic analyses based on nucleotide sequences of the partial glycoprotein and nucleoprotein genes of Lassa virus (LASV), showing the placement of the new sequences (in boldface) isolated from Mus baoulei pygmy mice, in comparison with other sequences representing the members of LASV lineages I–IV. A) Glycoprotein, 1,408 nt; B) nucleoprotein, 1,654 nt. The trees are rooted by Gbagroube, a LASV-like virus isolated from Mus setulosus mice in Côte d’Ivoire. Statistical support of grouping from Bayesian posterior probabilities is indicated at the nodes. Country, strain names, and GenBank accession numbers are indicated on the branches. The analysis was inferred by using the Bayesian Markov chain Monte Carlo method implemented in BEAST (8). The following settings were used: general time reversible plus gamma, strict clock, and constant population. Markov chain Monte Carlo chains were run for 10 million states and sampled every 10,000 states to obtain an effective sample size >200 for all parameters. The new viral and murine sequences are deposited under accession nos. MH028396–404. Scale bars indicate nucleotide substitutions per site.
References
- Whitmer SLM, Strecker T, Cadar D, Dienes HP, Faber K, Patel K, et al. New lineage of Lassa virus, Togo, 2016. Emerg Infect Dis. 2018;24:599–602. DOIPubMedGoogle Scholar
- World Health Organization. Lassa fever–Benin [cited 2016 July 1]. http://wwwwhoint/csr/don/13-june-2016-lassa-fever-benin
- Fichet-Calvet E. Lassa fever: a rodent-human interaction. In: Johnson N, editor. The role of animals in emerging viral diseases. London: Elsevier; 2014. p. 89–123.
- Kronmann KC, Nimo-Paintsil S, Guirguis F, Kronmann LC, Bonney K, Obiri-Danso K, et al. Two novel arenaviruses detected in pygmy mice, Ghana. Emerg Infect Dis. 2013;19:1832–5. DOIPubMedGoogle Scholar
- Olayemi A, Obadare A, Oyeyiola A, Fasogbon A, Igbokwe J, Igbahenah F, et al. Small mammal diversity and dynamics within Nigeria, with emphasis on reservoirs of the Lassa virus. Syst Biodivers. 2017;15:1–10.
- Olschläger S, Lelke M, Emmerich P, Panning M, Drosten C, Hass M, et al. Improved detection of Lassa virus by reverse transcription-PCR targeting the 5′ region of S RNA. J Clin Microbiol. 2010;48:2009–13. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, et al. RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Bowen MD, Rollin PE, Ksiazek TG, Hustad HL, Bausch DG, Demby AH, et al. Genetic diversity among Lassa virus strains. J Virol. 2000;74:6992–7004. DOIPubMedGoogle Scholar
- Olayemi A, Cadar D, Magassouba N, Obadare A, Kourouma F, Oyeyiola A, et al. New hosts of the Lassa virus. Sci Rep. 2016;6:25280. DOIPubMedGoogle Scholar