Volume 25, Number 3—March 2019
Research
Use of Genomics to Investigate Historical Importation of Shiga Toxin–Producing Escherichia coli Serogroup O26 and Nontoxigenic Variants into New Zealand
Figure 1
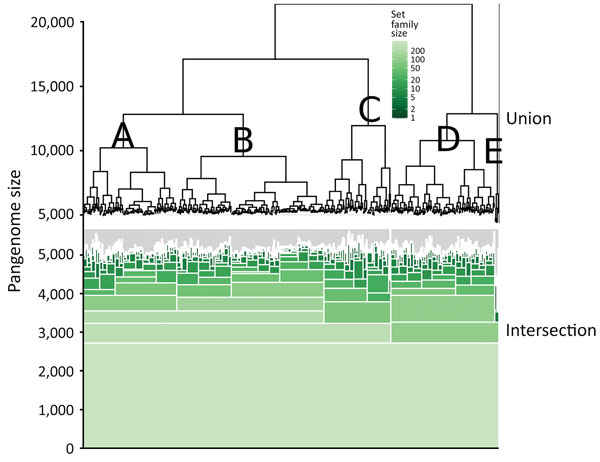
Figure 1. Hierarchical set analysis of 404 Escherichia coli serogroup O26 isolates in investigation of historical importation of Shiga toxin–producing E. coli serogroup O26 and nontoxigenic variants into New Zealand, with a hierarchical set RaxML pangenome tree (top of figure) and shared gene groups visualized in green (bottom of figure). This figure illustrates shared gene groups after pangenome analysis. The union portion represents the pangenome relatedness between bacterial isolates. A–E indicate clades.
1Current affiliation: Centers for Disease Control and Prevention, Christiansted, Virgin Islands, USA.
Page created: February 19, 2019
Page updated: February 19, 2019
Page reviewed: February 19, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.