Volume 26, Number 3—March 2020
Research
Multidrug- and Extensively Drug-Resistant Mycobacterium tuberculosis Beijing Clades, Ukraine, 2015
Figure 2
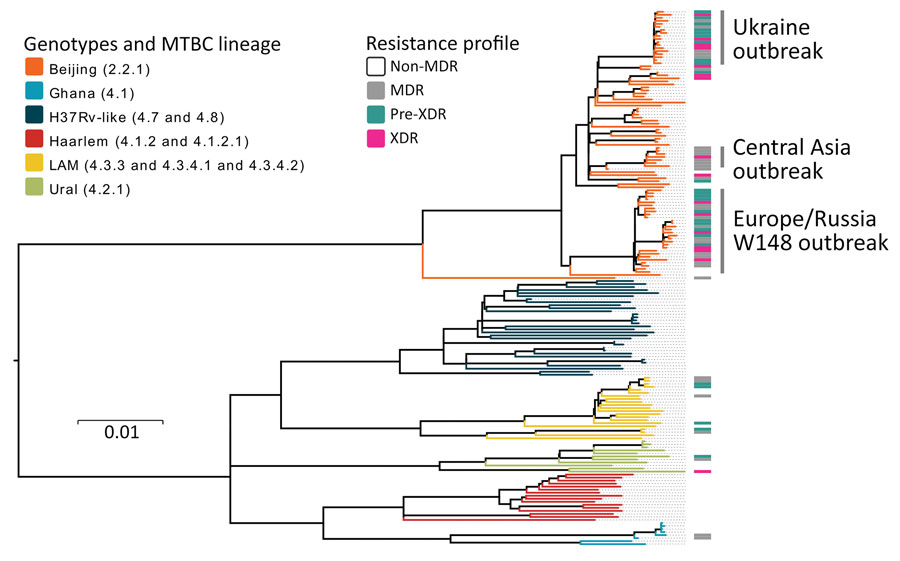
Figure 2. Maximum-likelihood phylogeny based on 10,339 SNPs, and employing general time-reversible substitution model for 177 clinical MDR/XDR and non-MDR Mycobacterium tuberculosis complex isolates from southern Ukraine. Branches are color-coded according to the phylogenetic classification from Coll et al. (22). Resistance profile bars represent drug resistance classifications based on drug resistance mediating mutations. Scale bar indicates substitutions per site. MDR, multidrug resistant; XDR, extensively drug-resistant.
References
- World Health Organization. Global tuberculosis report 2017. Geneva: The Organization; 2017.
- World Health Organization. Global tuberculosis report 2018: Geneva: The Organization; 2018.
- Walker TM, Merker M, Knoblauch AM, Helbling P, Schoch OD, van der Werf MJ, et al.; MDR-TB Cluster Consortium. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: a molecular epidemiological study. Lancet Infect Dis. 2018;18:431–40. DOIPubMedGoogle Scholar
- Lange C, Abubakar I, Alffenaar JW, Bothamley G, Caminero JA, Carvalho AC, et al.; TBNET. Management of patients with multidrug-resistant/extensively drug-resistant tuberculosis in Europe: a TBNET consensus statement. Eur Respir J. 2014;44:23–63. DOIPubMedGoogle Scholar
- Günther G, Gomez GB, Lange C, Rupert S, van Leth F; TBNET. Availability, price and affordability of anti-tuberculosis drugs in Europe: a TBNET survey. Eur Respir J. 2015;45:1081–8. DOIPubMedGoogle Scholar
- Matteelli A, Rendon A, Tiberi S, Al-Abri S, Voniatis C, Carvalho ACC, et al. Tuberculosis elimination: where are we now? Eur Respir Rev. 2018;27:27. DOIPubMedGoogle Scholar
- European Centre for Disease Prevention and Control/WHO Regional Office for Europe. Tuberculosis surveillance and monitoring in Europe, 2018, 2016 data. 2018 [cited 2019 Dec 31]. https://www.ecdc.europa.eu/en/publications-data/tuberculosis-surveillance-and-monitoring-europe-2018
- Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47:242–9. DOIPubMedGoogle Scholar
- Cohen KA, Abeel T, Manson McGuire A, Desjardins CA, Munsamy V, Shea TP, et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole-genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015;12:
e1001880 . DOIPubMedGoogle Scholar - Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–86. DOIPubMedGoogle Scholar
- Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–82. DOIPubMedGoogle Scholar
- Kendall EA, Fofana MO, Dowdy DW. Burden of transmitted multidrug resistance in epidemics of tuberculosis: a transmission modelling analysis. Lancet Respir Med. 2015;3:963–72. DOIPubMedGoogle Scholar
- Eldholm V, Monteserin J, Rieux A, Lopez B, Sobkowiak B, Ritacco V, et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015;6:7119. DOIPubMedGoogle Scholar
- Lalor MK, Casali N, Walker TM, Anderson LF, Davidson JA, Ratna N, et al. The use of whole-genome sequencing in cluster investigation of a multidrug-resistant tuberculosis outbreak. Eur Respir J. 2018;51:
1702313 . DOIPubMedGoogle Scholar - Nikolayevskyy V, Kranzer K, Niemann S, Drobniewski F. Whole genome sequencing of Mycobacterium tuberculosis for detection of recent transmission and tracing outbreaks: A systematic review. Tuberculosis (Edinb). 2016;98:77–85.https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=27156621&dopt=Abstract DOIGoogle Scholar
- Public Health Center Ukraine. Tuberculosis in Ukraine [in Ukrainian]. 2018 [cited 2019 Dec 31]. http://aph.org.ua/wp-content/uploads/2018/09/proekt-dovidnika-TB-2018.pdf
- van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–86. DOIPubMedGoogle Scholar
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. DOIPubMedGoogle Scholar
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. DOIPubMedGoogle Scholar
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. DOIPubMedGoogle Scholar
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:
e9490 . DOIPubMedGoogle Scholar - Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. DOIPubMedGoogle Scholar
- Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–46. DOIPubMedGoogle Scholar
- Coll F, McNerney R, Preston MD, Guerra-Assunção JA, Warry A, Hill-Cawthorne G, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015;7:51. DOIPubMedGoogle Scholar
- Feuerriegel S, Köser CU, Niemann S. Phylogenetic polymorphisms in antibiotic resistance genes of the Mycobacterium tuberculosis complex. J Antimicrob Chemother. 2014;69:1205–10. DOIPubMedGoogle Scholar
- Mayer D, Zeileis AKH. Visualizing independence using extended association plots. Presented at: Third International Workshop on Distributed Statistical Computing (DSC 2003); March 20–22, 2003; Vienna, Austria.
- The R Core Team. R: a language and environment for statistical computing. Reference index. 2019 [cited 2019 Dec 31]. https://cran.r-project.org/manuals.html
- Daum LT, Konstantynovska OS, Solodiankin OS, Liashenko OO, Poteiko PI, Bolotin VI, et al. Next-generation sequencing for characterizing drug resistance-conferring Mycobacterium tuberculosis genes from clinical isolates in the Ukraine. J Clin Microbiol. 2018;56:e00009–00018. DOIPubMedGoogle Scholar
- Nikolayevskyy VV, Brown TJ, Bazhora YI, Asmolov AA, Balabanova YM, Drobniewski FA. Molecular epidemiology and prevalence of mutations conferring rifampicin and isoniazid resistance in Mycobacterium tuberculosis strains from the southern Ukraine. Clin Microbiol Infect. 2007;13:129–38. DOIPubMedGoogle Scholar
- World Health Organization. Review of the national tuberculosis programme in Ukraine. Copenhagen, Denmark: The Organization; 2011.
- A unified clinical protocol of primary, secondary (specialized) and tertiary (highly specialized) medical care for adults [in Ukrainian]. Tuberculosis. 2014 [cited 2020 Jan 8]. https://www.phc.org.ua/sites/default/files/uploads/files/%D0%9F%D1%80%D0%BE%D1%82%D0%BE%D0%BA%D0%BE%D0%BB%20%D0%A2%D0%91%20%D1%83%20%D0%B2%D0%B7%D1%80%D0%BE%D1%81%D0%BB%D1%8B%D1%85%20620%20%D0%BE%D1%82%20%2004%2009%2014.pdf
- Nikolayevskyy V, Kontsevaya I, Nikolaevskaya E, Surkova E, Samchenko S, Esipenko S. Diagnostic performance and impact of routinely implemented Xpert® MTB/RIF assay in a setting of high incidence of drug-resistant TB in Odessa Oblast, Ukraine. Clin Microbiol Infect. 2019;25:1040.e1–6. DOIPubMedGoogle Scholar
- Manson AL, Cohen KA, Abeel T, Desjardins CA, Armstrong DT, Barry CE III, et al.; TBResist Global Genome Consortium. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat Genet. 2017;49:395–402. DOIPubMedGoogle Scholar
- Lange C, Duarte R, Fréchet-Jachym M, Guenther G, Guglielmetti L, Olaru ID, et al.; European MDR-TB database collaboration *. European MDR-TB database collaboration. Limited benefit of the new shorter multidrug-resistant tuberculosis regimen in Europe. Am J Respir Crit Care Med. 2016;194:1029–31. DOIPubMedGoogle Scholar
- Balabanova Y, Fiebig L, Ignatyeva O, Riekstina V, Danilovits M, Jaama K, et al. Multidrug-resistant TB in Eastern region of the EU: is the shorter regimen an exception or a rule? Thorax. 2017;72:850–2. DOIPubMedGoogle Scholar
- Konstantynovska O, Rekrotchuk M, Hrek I, Rohozhyn A, Rudova N, Poteiko P, et al. Severe clinical outcomes of tuberculosis in Kharkiv Region, Ukraine, are associated with Beijing strains of Mycobacterium tuberculosis. Pathogens. 2019;8:8. DOIPubMedGoogle Scholar
- Pavlenko E, Barbova A, Hovhannesyan A, Tsenilova Z, Slavuckij A, Shcherbak-Verlan B, et al. Alarming levels of multidrug-resistant tuberculosis in Ukraine: results from the first national survey. Int J Tuberc Lung Dis. 2018;22:197–205. DOIPubMedGoogle Scholar
- Hoffmann H, Kohl TA, Hofmann-Thiel S, Merker M, Beckert P, Jaton K, et al. Delamanid and bedaquiline resistance in Mycobacterium tuberculosis ancestral Beijing genotype causing extensively drug-resistant tuberculosis in a Tibetan refugee. Am J Respir Crit Care Med. 2016;193:337–40. DOIPubMedGoogle Scholar
Page created: February 20, 2020
Page updated: February 20, 2020
Page reviewed: February 20, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.