Volume 27, Number 12—December 2021
Research
Novel Filoviruses, Hantavirus, and Rhabdovirus in Freshwater Fish, Switzerland, 2017
Figure 2
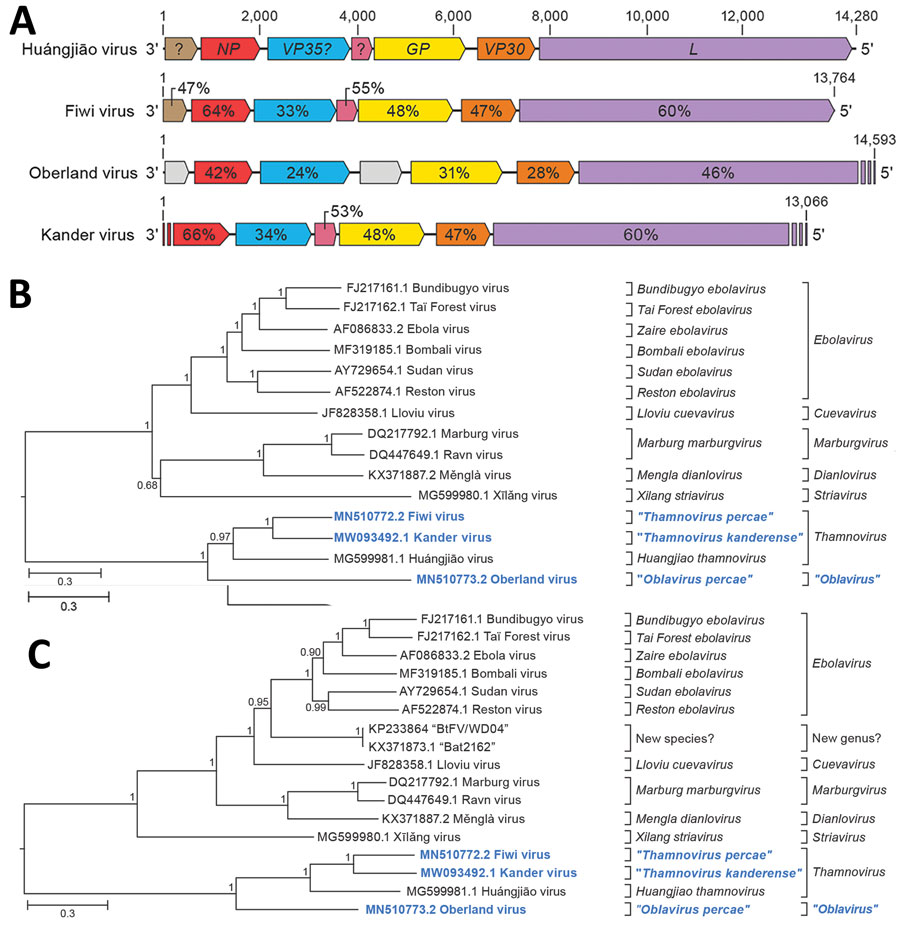
Figure 2. Identifying 3 novel filoviruses in European perch. A) Schematic representation of the genome organization of Fiwi virus, Oberland virus, and Kander virus compared with Huángjiāo virus (HUJV). Open reading frames (ORFs) are indicated by colored arrows. ORFs encoding HUJV-like proteins are depicted by the same color and sequence similarities are indicated as percentages. Undetermined ORF starts and ends are shown as stripes. B, C) Maximum-likelihood phylogenetic trees of the new filovirus genome sequences (bold blue) generated by using coding-complete and near-complete genome sequences (B) or only L gene sequences (C) of representative members of the family Filoviridae. Numbers near nodes on the trees indicate bootstrap values. Branches are labeled by GenBank accession number, and virus name. Scale bar indicates number of substitutions per site, reflected by branch lengths. GP, glycoprotein gene; L, large protein gene; NP, nucleoprotein gene; VP30, transcriptional activator gene; VP35, polymerase cofactor gene.
1These authors contributed equally to this article.