Volume 27, Number 4—April 2021
Dispatch
Genomic Analysis of Novel Poxvirus Brazilian Porcupinepox Virus, Brazil, 2019
Figure 2
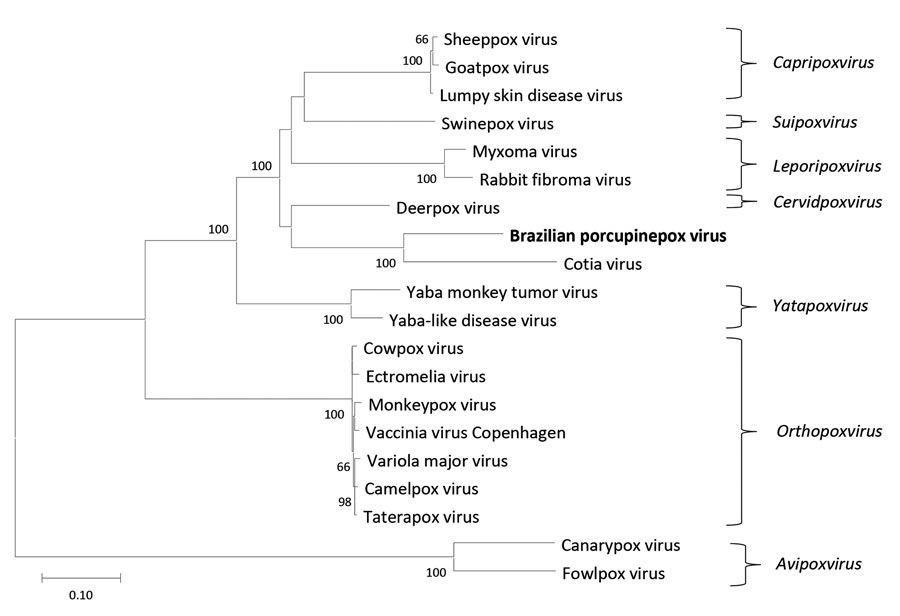
Figure 2. Phylogenetic tree constructed in genomic analysis of novel poxvirus Brazilian porcupinepox virus, Brazil, 2019 (boldface). Tree constructed by using the maximum-likelihood method and Jones–Taylor–Thornton model (7) with frequency model for amino acid sequence alignments of the RNA polymerase subunit RPO147, RNA polymerase subunit RPO132, RNA polymerase–associated RAP94, mRNA capping enzyme large subunit, virion major core protein P4a, early transcription factor VETFL, nucleoside-triphosphatase, DNA polymerase, and DNA topoisomerase I genes of selected strains representing different genera of chordopoxvirus with low GC contents and their respective genera. The numbers next to each node represent the values of 1,000 bootstrap repetitions, and only those >50% are shown. Evolutionary analyses were conducted in MEGA X (8). GenBank accession numbers are as follows: Brazilian porcupinepox virus, MK944278.1; camelpox virus, AY009089.1; canarypox virus, NC005309.1; Cotia virus, KM595078.1; cowpox virus, DQ437593.1; deerpox virus, AY689437.1; ectromelia virus, NC004105.1; fowlpox virus, NC002188.1; goatpox virus, MH381810.1; lumpy skin disease virus, NC003027.1; monkeypox virus, DQ011157.1; myxoma virus, NC001132.2; rabbit fibroma virus, NC001266.1; sheeppox virus, NC004002.1; swinepox virus, NC003389.1; taterapox virus, NC008291.1; vaccinia virus, M35027.1; variola major virus, L22579.1; Yaba monkey tumor virus, NC005179.1; Yaba-like disease virus, NC002642.1. Scale bar represents number of substitutions per site.
References
- Oliveira GP, Rodrigues RAL, Lima MT, Drumond BP, Abrahão JS. Poxvirus host range genes and virus–host spectrum: a critical review. Viruses. 2017;9:331. DOIPubMedGoogle Scholar
- Gigante CM, Gao J, Tang S, McCollum AM, Wilkins K, Reynolds MG, et al. Genome of Alaskapox virus, a novel orthopoxvirus isolated from Alaska. Viruses. 2019;11:708. DOIPubMedGoogle Scholar
- Kantele A, Chickering K, Vapalahti O, Rimoin AW. Emerging diseases-the monkeypox epidemic in the Democratic Republic of the Congo. Clin Microbiol Infect. 2016;22:658–9. DOIPubMedGoogle Scholar
- Abrahão JS, Campos RK, Trindade GS, Guimarães da Fonseca F, Ferreira PCP, Kroon EG. Outbreak of severe zoonotic vaccinia virus infection, Southeastern Brazil. Emerg Infect Dis. 2015;21:695–8. DOIPubMedGoogle Scholar
- Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–76. DOIPubMedGoogle Scholar
- Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol. 2003;77:7590–600. DOIPubMedGoogle Scholar
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Lopesode S, Lacerda JP, Fonseca IE, Castro DP, Forattini OP, Rabello EX. Cotia virus: a new agent isolated from sentinel mice in São Paulo, Brazil. Am J Trop Med Hyg. 1965;14:156–7. DOIPubMedGoogle Scholar
- Afonso PP, Silva PM, Schnellrath LC, Jesus DM, Hu J, Yang Y, et al. Biological characterization and next-generation genome sequencing of the unclassified Cotia virus SPAn232 (Poxviridae). J Virol. 2012;86:5039–54. DOIPubMedGoogle Scholar
- International Committee on Taxonomy of Viruses. Poxviridae. 2011 [cited 2020 Jul 22]. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/dsdna-viruses-2011/w/dsdna_viruses/74/poxviridae
- Marinho-Filho J, Emmons L. Coendou prehensilis (Brazilian porcupine). The IUCN red list of threatened species. 2016 [cited 2020 Oct 29]. https://www.iucnredlist.org/species/101228458/22214580
- Emmons LH, Feer F. Porcupines (Erethizontidae). In: Emmons LH, Feer F, editors. Neotropical rainforest mammals: a field guide. 2nd ed. Chicago: University of Chicago Press; 1997. p. 216–23.
- Essbauer S, Pfeffer M, Meyer H. Zoonotic poxviruses. Vet Microbiol. 2010;140:229–36. DOIPubMedGoogle Scholar
- Barrett JW, McFadden G. Origin and evolution of poxviruses. In: Domingo E, Parrish C, Holland J, editors. Origin and evolution of viruses. 2nd ed. Cambridge (MA): Academic Press; 2008. p. 431–46.