Volume 27, Number 4—April 2021
Dispatch
Genomic Analysis of Novel Poxvirus Brazilian Porcupinepox Virus, Brazil, 2019
Abstract
We obtained the complete sequence of a novel poxvirus, tentatively named Brazilian porcupinepox virus, from a wild porcupine (Coendou prehensilis) in Brazil that had skin and internal lesions characteristic of poxvirus infection. The impact of this lethal poxvirus on the survival of this species and its potential zoonotic importance remain to be investigated.
Poxviruses are among the best known and most feared viruses (1); the Poxviridae family includes several viruses of veterinary and medical relevance, some of them zoonotic. Emergence and reemergence of poxviruses is frequently observed (2–4). We report a systemic and lethal poxvirus infection in a wild porcupine and further characterize the virus through genomic analysis.
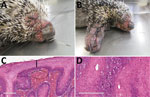
In March 2019, a free-ranging adult male Brazilian porcupine (Coendou prehensilis) in good bodily condition was captured near a park in the urban area of Uberlândia in the state of Minas Gerais in southeastern Brazil and was then referred for veterinary clinical care at the Federal University of Uberlândia. The animal had multifocal skin edema and erythema, especially on the eyelid and muzzle (Figure 1, panel A), extremity of limbs (Figure 1, panel B), and genital areas, and a penetrating skin lesion on the lateral face of the right limb near the elbow joint. We observed purulent nasal and ocular secretion. After 4 days of supportive treatment, the porcupine died and was subjected to a full necropsy for histopathologic evaluation and to collect samples for molecular investigation.
On macroscopic examination, the spleen was enlarged, lungs were turgid, and the liver was pale and greyish. Standard histopathologic sections were cut from formalin-fixed, paraffin-embedded skin and organ samples and stained with hematoxylin and eosin. Histologic examination of the skin revealed several common alterations, irrespective of the location of the sample (Figure 1, panels C and D). Epidermis exhibited marked hyperplasia, parakeratotic hyperkeratosis, and moderate acantholysis. Epidermal cells were swollen, with foci of ballooning degeneration, and the cytoplasm of scattered epithelial cells contained round eosinophilic inclusions of varying size. No intranuclear inclusions were found. We also observed ulcerated epidermis with eosinophilic, amorphous keratinaceous crusts, necrosis, and numerous degenerated granulocytes. Dermal lesions included hemorrhage at the dermal–epidermal junction, severe edema, necrotic areas, and mixed inflammatory infiltrate that extended into the deep dermis. Venous blood congestion was observed in the kidneys, liver, spleen, and lungs. Hepatocytes evidenced moderate degeneration, whereas emphysema and pneumonitis were observed in the lungs.
We extracted total DNA from lesioned eyelid skin, spleen, and liver samples and subjected to a pan-pox universal PCR assay (5). All samples resulted in amplicons with low–GC content poxvirus primers targeting a region of the putative metalloproteinase gene. The amplicon from lesioned eyelid skin was submitted for Sanger sequencing (GenBank accession no. MK944278), and total DNA from this lesion was submitted to full-genome sequencing using the Illumina NextSeq platform (Illumina, https://www.illumina.com). We performed viral particle enrichment and next-generation sequencing (Appendix). A total of 71,507,840 pairs of 151-bp reads were obtained after raw data quality control using CLC Genomics Workbench 11 (QIAGEN, https://www.qiagen.com). The resulting paired-end reads were de novo assembled in CLC Genomics Workbench 11 (QIAGEN) with default parameters, resulting in a 144,504-nt genome (GenBank accession no. MN692191) with average coverage of 230.41x.
We annotated the genome (Appendix). Of 133 open reading frames (ORFs) found, only 2 (117 and 129) have no equivalents in other poxviruses, ORFs situated in the middle region of the genome encode proteins related to virion morphogenesis, the structure of virus particles, and viral DNA and RNA metabolism. We also identified the minimum essential chordopoxvirus genome, 49 genes conserved between highly diverged poxviruses families, and 41 genes conserved in chordopoxviruses (6). ORFs related to host range, immunomodulation, and virulence were observed in the extremities of the genome.
Alignments of 9 amino acid sequences corresponding to 9 conserved genes located in the central region were concatenated (7,956 aa) compiled from this genome with homologous sequences from different genera of chordopoxviruses with low GC content. Thus, we constructed a phylogenetic tree by using the maximum-likelihood method and Jones–Taylor–Thornton model (7) in MEGA X software (8) with the frequency matrix model (Figure 2). A nucleotide tree with complete genomes was also constructed (Appendix Figure 1).
Using the 9 concatenated sequences corresponding to the conserved central region of MN692191 and other known chordopoxviruses, we obtained nucleotide identities ranged from 69.9% to 85.2% (Appendix Figure 2) and the amino acid identities ranged from 57.7% to 78.8% (Appendix Figure 3). The highest identity was observed between MN692191 and Cotia virus (CoTV) in clusters in the phylogenetic trees. In the 1960s, CoTV was isolated in Brazil from sentinel suckling mice (9); the natural host remains unknown and CoTV remains unclassified despite attempts to place it in a new genus of Poxviridae (10). According to the International Committee on Taxonomy of Viruses (11), isolates within a species exhibit >98% nucleotide identity. CoTV and MN692191 exhibit 85.2% of nucleotide identity and thus are distinct species.
We estimated maximum-likelihood distances for this region of conserved nucleotides; the distance between CoTV and MN692191 was 0.156. MN692191 belongs to a main clade that includes Capripoxvirus, Suipoxvirus, Leporipoxvirus, Cervidpoxvirus, and Yatapoxvirus (clade CSLCY). Distances ranging from 0.177 to 0.210 were observed between MN692191 and species from different genera of the CSLCY clade. In species from the same genus of this clade, distance ranged from 0.006 to 0.052, which suggests that MN692191 and CoTV (maximum-likelihood distance 0.156) do not belong to the same genus.
This comprehensive phylogenetic analysis supports the classification of MN692191 into a new genus in the family Poxviridae, subfamily Chordopoxvirinae. Because the virus described in this study is distinct from previously identified viruses, we propose the tentative species name Brazilian porcupinepox virus (BPoPV), according to recommendations for nomenclature of poxvirus species of the International Committee on Taxonomy of Viruses (11).
Wildlife veterinarians in Brazil have observed free-ranging porcupines exhibiting clinical signs compatible with those described in this study (B.S.S. Petri, CRAS Parque Ecológico do Tietê- São Paulo, pers. comm., 2019 Sep 16; I.S. Barbosa, CETAS–Goiânia, pers. comm., 2020 Jan 6) have been observed. In 2019, of 13 of these porcupine specimens reported, only 3 had fully recovered from clinical symptoms, demonstrating that this virus might be a common pathogen for this species and could have consequences for its conservation.
Brazilian porcupines have a greater distribution in Brazil but are found in 10 other countries in Latin America (12). This species is found mainly in forest environments (12) and can be observed in forest fragments in urban areas, as was the case for the specimen in this study. Housing construction nearer to forested areas has led to this porcupine sometimes being hunted for meat (13), which leads to human exposure to the pathogens hosted by this species.
The genus Orthopoxvirus includes the best-known zoonotic poxvirus species, such as cowpox, monkeypox, and vaccinia viruses (14); however, the zoonotic poxvirus is not restricted to this genus. The genera Parapoxvirus and Yatapoxvirus also include viral species of zoonotic importance (15). Furthermore, chordopoxviruses are often described as emerging zoonoses. Contact between Brazilian porcupines and humans, because of anthropized forested areas and the porcupines’ broad geographic distribution and presence in urban areas, raises concerns about the zoonotic potential of BPoPV, which remains to be investigated.
In summary, our description of this novel poxvirus contributes to knowledge of viral diversity and pathogenicity of poxviruses. Some chordopoxviruses are capable of infecting multiple animal species, whereas others have a restricted host spectrum (14,15). The infection capability of BPoPV is a crucial aspect for further study.
Dr. Hora is a professor in the School of Veterinary Medicine, Federal University of Uberlândia, Brazil. Her research interests include molecular diagnostic techniques of infectious diseases in wild and domestic animals.
Acknowledgments
We thank José Luiz Catão-Dias and Carlos Sacristán for kindly providing the positive control for the initial molecular screening.
This research was sponsored by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (finance code 001) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant no. 307291/2017-0).
References
- Oliveira GP, Rodrigues RAL, Lima MT, Drumond BP, Abrahão JS. Poxvirus host range genes and virus–host spectrum: a critical review. Viruses. 2017;9:331. DOIPubMedGoogle Scholar
- Gigante CM, Gao J, Tang S, McCollum AM, Wilkins K, Reynolds MG, et al. Genome of Alaskapox virus, a novel orthopoxvirus isolated from Alaska. Viruses. 2019;11:708. DOIPubMedGoogle Scholar
- Kantele A, Chickering K, Vapalahti O, Rimoin AW. Emerging diseases-the monkeypox epidemic in the Democratic Republic of the Congo. Clin Microbiol Infect. 2016;22:658–9. DOIPubMedGoogle Scholar
- Abrahão JS, Campos RK, Trindade GS, Guimarães da Fonseca F, Ferreira PCP, Kroon EG. Outbreak of severe zoonotic vaccinia virus infection, Southeastern Brazil. Emerg Infect Dis. 2015;21:695–8. DOIPubMedGoogle Scholar
- Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–76. DOIPubMedGoogle Scholar
- Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol. 2003;77:7590–600. DOIPubMedGoogle Scholar
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Lopesode S, Lacerda JP, Fonseca IE, Castro DP, Forattini OP, Rabello EX. Cotia virus: a new agent isolated from sentinel mice in São Paulo, Brazil. Am J Trop Med Hyg. 1965;14:156–7. DOIPubMedGoogle Scholar
- Afonso PP, Silva PM, Schnellrath LC, Jesus DM, Hu J, Yang Y, et al. Biological characterization and next-generation genome sequencing of the unclassified Cotia virus SPAn232 (Poxviridae). J Virol. 2012;86:5039–54. DOIPubMedGoogle Scholar
- International Committee on Taxonomy of Viruses. Poxviridae. 2011 [cited 2020 Jul 22]. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/dsdna-viruses-2011/w/dsdna_viruses/74/poxviridae
- Marinho-Filho J, Emmons L. Coendou prehensilis (Brazilian porcupine). The IUCN red list of threatened species. 2016 [cited 2020 Oct 29]. https://www.iucnredlist.org/species/101228458/22214580
- Emmons LH, Feer F. Porcupines (Erethizontidae). In: Emmons LH, Feer F, editors. Neotropical rainforest mammals: a field guide. 2nd ed. Chicago: University of Chicago Press; 1997. p. 216–23.
- Essbauer S, Pfeffer M, Meyer H. Zoonotic poxviruses. Vet Microbiol. 2010;140:229–36. DOIPubMedGoogle Scholar
- Barrett JW, McFadden G. Origin and evolution of poxviruses. In: Domingo E, Parrish C, Holland J, editors. Origin and evolution of viruses. 2nd ed. Cambridge (MA): Academic Press; 2008. p. 431–46.
Figures
Cite This ArticleOriginal Publication Date: March 09, 2021
Table of Contents – Volume 27, Number 4—April 2021
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
A.S. Hora, Avenida Ceará S/N, bloco 2D, sala 46, Campus Umuarama, Uberlândia–Minas Gerais, 38400-902, Brazil
Top