Volume 27, Number 6—June 2021
Dispatch
Serotype-Switch Variant of Multidrug-Resistant Streptococcus pneumoniae Sequence Type 271
Figure
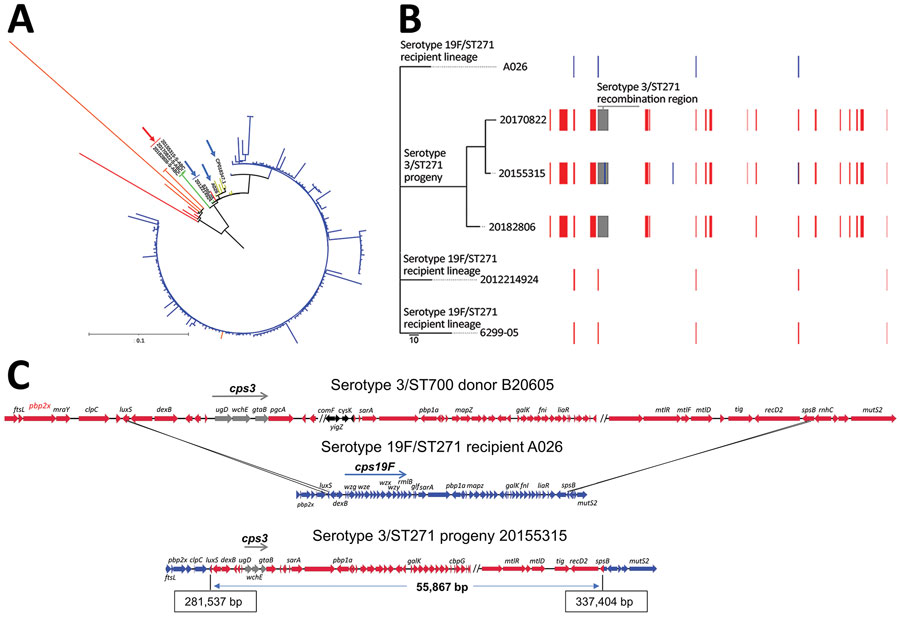
Figure. Streptococcus pneumoniae serotype 3/ST271 lineage resulting from a recombination event between a 19F/ST271 recipient and 3/ST700 donor. A) Phylogenetic tree showing progeny serotype 3/ST271 isolates 20155315-S-ABC, 20170822-S-ABC, and 20182806-S-ABC (red arrow) most closely related to the putative recipient 19F/ST271 isolates 6299-05 and 2012214924 (blue arrows). The most closely related ST271 single contig reference is A026 (also indicated with blue arrow). Branch colors: yellow, additional 19F/ST271 Active Bacterial Core surveillance (ABCs) isolates and single contig references; red, single-locus variant single contig references or ABCs isolates; orange, double-locus variant single contig reference or ABCs isolates; blue, ST320 ABCs isolates. Zero-, single-, and double-locus variant, single contig references were identified by using the PubMLST database (https://pubmlst.org). Scale bar corresponds to 1,062 single nucleotide polymorphisms. B) Phylogenetic alignment of the 3 recombinant serotype 3/ST271 isolates and closest known genomic matches of the ST271 recipient lineage and a schematic of recombinant genome fragments, represented by rectangular blocks, that were predicted by Gubbins (10). Block locations and sizes are relative to the aligned genomes; red blocks represent sites in common between >2 isolates (1 site, <150 bp in length, was not counted among the 17 total shown), blue blocks sites unique to a given isolate, and gray blocks the serotype-switch fragment that replaced the corresponding cps19F region within the recipient 19F/ST271 strain. The cps3 locus was not identified using Gubbins because of its complete divergence from cps19F and instead was identified using ProgressiveMauve (12) within the encompassed 55.9 kb fragment (panel C). Gray block contains cps3, pbp1a (PBP1A-17), trigger factor, and choline binding protein G genes. C) Schematic illustrating ancestral recombination event between the 3/ST700 donor (B20605) and 19F/ST271 recipient (A026) to yield the 3/ST271 progeny (20155315-S-ABC). Deduced crossover points, including coordinates in the progeny, are shown. luxS and spsB genes are shown as blue/red hybrids. The minimum genes of the cps3 operon required for polysaccharide capsuale biosynthesire shown in gray (ugd, wchE, gtaB). Genes with arrows in black differ between B20605 and 73D36881 and are absent in the 3/ST271 isolates. ST, sequence type.
References
- Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, et al.; Active Bacterial Core Surveillance/Emerging Infections Program Network. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. 2010;201:32–41. DOIPubMedGoogle Scholar
- Moore MR, Link-Gelles R, Schaffner W, Lynfield R, Lexau C, Bennett NM, et al. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: analysis of multisite, population-based surveillance. Lancet Infect Dis. 2015;15:301–9. DOIPubMedGoogle Scholar
- Beall BW, Gertz RE, Hulkower RL, Whitney CG, Moore MR, Brueggemann AB. Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J Infect Dis. 2011;203:1360–8. DOIPubMedGoogle Scholar
- Beall B, Chochua S, Gertz RE Jr, Li Y, Li Z, McGee L, et al. A population-based descriptive atlas of invasive pneumococcal strains recovered within the U.S. during 2015–2016. Front Microbiol. 2018;9:2670. DOIPubMedGoogle Scholar
- Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:
e31 . DOIPubMedGoogle Scholar - Poolman J, Kriz P, Feron C, Di-Paolo E, Henckaerts I, Miseur A, et al. Pneumococcal serotype 3 otitis media, limited effect of polysaccharide conjugate immunisation and strain characteristics. Vaccine. 2009;27:3213–22. DOIPubMedGoogle Scholar
- Choi EH, Zhang F, Lu YJ, Malley R. Capsular polysaccharide (CPS) release by serotype 3 pneumococcal strains reduces the protective effect of anti-type 3 CPS antibodies. Clin Vaccine Immunol. 2015;23:162–7. DOIPubMedGoogle Scholar
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. DOIPubMedGoogle Scholar
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. DOIPubMedGoogle Scholar
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:
e11147 . DOIPubMedGoogle Scholar - Kilian M, Tettelin H. Identification of virulence-associated properties by comparative genome analysis of Streptococcus pneumoniae, S. pseudopneumoniae, S. mitis, three S. oralis subspecies, and S. infantis. MBio. 2019;10:e01985–19. DOIPubMedGoogle Scholar
- Mann B, Orihuela C, Antikainen J, Gao G, Sublett J, Korhonen TK, et al. Multifunctional role of choline binding protein G in pneumococcal pathogenesis. Infect Immun. 2006;74:821–9. DOIPubMedGoogle Scholar
- Cohen A, Troib S, Dotan S, Najmuldeen H, Yesilkaya H, Kushnir T, et al. Streptococcus pneumoniae cell wall-localized trigger factor elicits a protective immune response and contributes to bacterial adhesion to the host. Sci Rep. 2019;9:4295. DOIPubMedGoogle Scholar
1Current affiliation: Emory University, Atlanta, Georgia, USA.