Volume 29, Number 1—January 2023
Dispatch
Survey of West Nile and Banzi Viruses in Mosquitoes, South Africa, 2011–2018
Abstract
We collected >40,000 mosquitoes from 5 provinces in South Africa during 2011–2018 and screened for zoonotic flaviviruses. We detected West Nile virus in mosquitoes from conservation and periurban sites and potential new mosquito vectors; Banzi virus was rare. Our results suggest flavivirus transmission risks are increasing in South Africa.
Flaviviruses have been major emerging zoonotic pathogens in Africa within the past decade (1). In South Africa, West Nile virus (WNV) is the main flavivirus detected in animals and humans (2,3). Several other lesser-known flaviviruses were first described in South Africa but are understudied and potentially underreported, including Wesselsbron, Usutu, and Banzi (BANV) viruses (4). In South Africa, mosquito surveillance is not routinely performed and studies on flavivirus ecology are outdated (5). In this study, we aimed to update flavivirus vector epidemiology in northeastern provinces of South Africa through a large-scale ecologic survey.
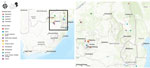
We selected 15 sites (4 sentinel sites, 11 ad hoc sites) across 5 provinces in South Africa for mosquito collection according to recent cases of arboviral disease in humans and animals (2,3) (Figure 1). We established sentinel sites in Boschkop and Kyalami, both located in the Gauteng province (periurban sites), and Lapalala and Marakele, both located in the Limpopo province (conservation sites); collections were performed during 2011–2018 (Table 1). Opportunistic supplementary collections occurred at 10 ad hoc sites during 2015–2018 spanning 3 additional provinces that included urban, periurban, and conservation sites. We performed additional ad hoc collections during March–April 2017 in and around Kruger National Park (KNP) located within Limpopo and Mpumalanga provinces (periurban/conservation sites) (6). The methodologies for mosquito collection, identification, pooling, processing, flavivirus screening, and COX1 gene sequencing have been previously described (7). To focus on months from midsummer to autumn in South Africa, when availability of mosquito breeding sites and vectorial capacity increases because of warmer and wetter weather conditions (8), we only screened mosquitoes collected during January–June. To submit sequences to GenBank, we generated NS5 gene fragments >200 bp by using heminested PCR with 0.4 µmol/L each of forward primer (FU1, 5′-TACAACATGGGAAAGAGAGAA-3′) and reverse primer (CFD2, 5′-GTGTCCCAGCCGGCGGTGTCATCAGC-3′) and Platinum Taq DNA Polymerase (Thermo Fisher Scientific, https://www.thermofisher.com). For WNV positive pools, we performed reverse transcription PCR to amplify a 1,525 bp fragment of the WNV envelope protein gene for phylogenetic analysis (Appendix Table). We calculated the mosquito minimum infection rate (MIR) per site by using a standard formula: (number of positive pools/total number of individual mosquitoes tested) × 1000.
We collected a total of 66,299 mosquitoes belonging to 11 genera across 5 provinces in South Africa during 2011–2018 (Table 1; Appendix Figure 1). From those, we divided 40,731 female mosquitoes into 1,471 pools based on morphology, focusing on only 4 genera (Culex, Mansonia, Anopheles, and Aedes) and screened for flaviviruses. We detected WNV in 16 (1.09%) and BANV in 2 (0.14%) pools. We did not detect other zoonotic flaviviruses; however, insect-specific flaviviruses were detected and described elsewhere (7). WNV outbreaks can be expected once the MIR rises above 1 (9). We observed the highest MIRs in Kyalami (periurban site, MIR = 2.53), KNP (periurban/conservation site, MIR = 1.22), and Lapalala (conservation site, MIR = 1.01) (Table 2). Therefore, we identified those areas as higher risk sites for WNV outbreaks in humans and animals. These results correlated with areas where Culex spp. mosquitoes were the most abundant and where WNV cases were previously reported in humans and animals in South Africa (3,10).
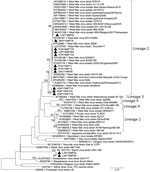
Only 11 of 16 WNV-positive pools had partial NS5 gene sequences of sufficient quality to perform maximum-likelihood analysis; we confirmed all 11 pools were WNV and also confirmed 2 BANV-positive pools (Figure 2; bootstrap value = 100 for both viruses). We observed high nucleotide similarity (94.62%–100.00%) between the identified WNV NS5 gene sequences and those from previously identified, highly neuroinvasive strains from South Africa isolated from either equines or humans. We successfully amplified the 1,525 nt region of the envelope protein gene for 5 of 16 WNV-positive pools (Appendix Figure 2).
We performed COX1 gene sequencing for 9 of 16 WNV-positive pools and confirmed morphologic identification of those mosquitoes as Cx. univittatus except for 1 pool collected in KNP that was identified by sequencing as Cx. perexiguus (Table 2; Appendix Figure 3), a mosquito species not known to be present in South Africa (11). This 1 pool might have contained a mix of both species, but Sanger sequencing was unable to distinguish between the 2 species. In addition, the COX1 reference sequences for Cx. perexiguus mosquitoes obtained from online databases may not be accurate because this species has not been identified in South Africa. Recently, COX1 gene amplification using universal primers followed by next-generation sequencing was shown to distinguish between species in mixed pools (12) and might be useful in future studies to resolve this ambiguity. Further studies are necessary to clarify the status of the Cx. perexiguus mosquitoes in South Africa. Most of the WNV-positive pools consisted of Cx. univittatus, Cx. pipiens s.l., and Cx. theileri mosquitoes, which we collected in high abundance. This result reiterates the importance of these species as WNV vectors in South Africa (5). We identified Cx. simpsoni, Cx. bitaeniorhynchus, An. gambiae sensu lato, and Cx. poicilipes mosquitoes, none of which have been previously associated with WNV in South Africa, as new potential vectors for WNV by using COX1 gene sequencing (4). Globally, from this list of species, only Cx. poicilipes mosquitoes from Senegal were found to be infected with WNV (13). Experimental studies have shown that the Cx. bitaeniorhynchus mosquito is a likely vector for WNV because this species was able to successfully transmit WNV (14). We were unable to genetically characterize the remaining 7 of 16 pools because of insufficient material for DNA extraction; we identified mosquitoes in those pools by morphologic characteristics.
Detection of BANV in mosquito pools in South Africa has not been described since the late 1970s (15), and a lack of surveillance raises the question regarding the true incidence of this virus. Only the Cx. rubinotus mosquito is recognized as a vector for BANV (15). Despite unclear morphologic identification, we identified Cx. rubinotus and Cx. annulioris mosquitoes in the 2 BANV-positive pools through COX1 gene sequencing (Table 2; Appendix Figure 3), which should be investigated further to confirm vector status.
The first limitation of our study is that we did not separate voucher specimens for mosquito species from the pools before homogenization. A voucher specimen is a mosquito species that is preserved and serves as a reference used to document identity. Second, identifications of mosquito species not previously associated with WNV infection are preliminary findings, and further investigation of vector competency is required.
Mosquito surveillance is not routinely performed for arboviruses in South Africa, and most studies were performed 40 years ago (5). In this study, 5 provinces were targeted for mosquito surveillance over a 7-year period. These investigations revealed a wide range of new potential vectors that require further investigation. Both WNV and BANV were identified in mosquitoes in periurban and conservation areas at the animal/human interface in South Africa, suggesting increasing circulation potential for those viruses between humans, wildlife, domestic animals, and avian species that are common in those areas.
Ms. MacIntyre is an MSc candidate in the Zoonotic Arbo- and Respiratory virus research group, Department of Medical Virology, at the University of Pretoria. Her research focuses on One Health investigations of West Nile virus in South Africa.
Acknowledgments
We thank Lapalala Wilderness, Marataba Conservation, and South African National Parks for logistical assistance and permission to collect mosquitoes; Basil Brooke and Danny Govender for their contribution to the collection of the mosquitoes; all previous staff and students in the Zoonotic Arbo- and Respiratory virus program for their technical support; and Clarence Yah for his assistance in preparing the manuscript. A.P.G. Almeida has been a recipient of the Visiting Professor Programme of the University of Pretoria and acknowledges the Global Health and Tropical Medicine unit.
The study was funded in part by a cooperative agreement (no. 5 NU2GGH001874-02-00) with the US Centers for Disease Control and Prevention. The study was solely the responsibility of the authors and does not necessarily represent the official views of the US Centers for Disease Control and Prevention or US Department of Health and Human Services.
References
- Hollidge BS, González-Scarano F, Soldan SS. Arboviral encephalitides: transmission, emergence, and pathogenesis. J Neuroimmune Pharmacol. 2010;5:428–42. DOIPubMedGoogle Scholar
- Burt FJ, Grobbelaar AA, Leman PA, Anthony FS, Gibson GV, Swanepoel R. Phylogenetic relationships of southern African West Nile virus isolates. Emerg Infect Dis. 2002;8:820–6. DOIPubMedGoogle Scholar
- Venter M, Pretorius M, Fuller JA, Botha E, Rakgotho M, Stivaktas V, et al. West Nile virus lineage 2 in horses and other animals with neurologic disease, South Africa, 2008–2015. Emerg Infect Dis. 2017;23:2060–4. DOIPubMedGoogle Scholar
- Venter M. Assessing the zoonotic potential of arboviruses of African origin. Curr Opin Virol. 2018;28:74–84. DOIPubMedGoogle Scholar
- Jupp PG. The ecology of West Nile virus in South Africa and the occurrence of outbreaks in humans. Ann N Y Acad Sci. 2001;951:143–52. DOIPubMedGoogle Scholar
- Gorsich EE, Beechler BR, van Bodegom PM, Govender D, Guarido MM, Venter M, et al. A comparative assessment of adult mosquito trapping methods to estimate spatial patterns of abundance and community composition in southern Africa. Parasit Vectors. 2019;12:462. DOIPubMedGoogle Scholar
- Guarido MM, Govender K, Riddin MA, Schrama M, Gorsich EE, Brooke BD, et al. Detection of insect-specific flaviviruses in mosquitoes (Diptera: Culicidae) in northeastern regions of South Africa. Viruses. 2021;13:2148. DOIPubMedGoogle Scholar
- Cornel AJ, Lee Y, Almeida APG, Johnson T, Mouatcho J, Venter M, et al. Mosquito community composition in South Africa and some neighboring countries. Parasit Vectors. 2018;11:331. DOIPubMedGoogle Scholar
- Bustamante DM, Lord CC. Sources of error in the estimation of mosquito infection rates used to assess risk of arbovirus transmission. Am J Trop Med Hyg. 2010;82:1172–84. DOIPubMedGoogle Scholar
- Zaayman D, Venter M. West Nile virus neurologic disease in humans, South Africa, September 2008-may 2009. Emerg Infect Dis. 2012;18:2051–4. DOIPubMedGoogle Scholar
- Mixão V, Bravo Barriga D, Parreira R, Novo MT, Sousa CA, Frontera E, et al. Comparative morphological and molecular analysis confirms the presence of the West Nile virus mosquito vector, Culex univittatus, in the Iberian Peninsula. Parasit Vectors. 2016;9:601. DOIPubMedGoogle Scholar
- Alkan C, Erisoz Kasap O, Alten B, de Lamballerie X, Charrel RN. Sandfly-borne phlebovirus isolations from Turkey: new insight into the sandfly fever Sicilian and sandfly fever Naples species. PLoS Negl Trop Dis. 2016;10:
e0004519 . DOIPubMedGoogle Scholar - Hubálek Z, Halouzka J. West Nile fever—a reemerging mosquito-borne viral disease in Europe. Emerg Infect Dis. 1999;5:643–50. DOIPubMedGoogle Scholar
- llkal MA, Mavale MS, Prasanna Y, Jacob PG, Geevarghese G, Banerjee K. Experimental studies on the vector potential of certain Culex species to West Nile virus. Indian J Med Res. 1997;106:225–8.
- Jupp PG, McIntosh BM, Anderson D. Culex (Eumelanomyia) rubinotus Theobald as vector of Banzi, Germiston and Witwatersrand viruses. IV. Observations on the biology of C. rubinotus. J Med Entomol. 1976;12:647–51. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: December 15, 2022
Table of Contents – Volume 29, Number 1—January 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Marietjie Venter, Zoonotic Arbo- and Respiratory Virus Program, Centre for Viral Zoonosis, Department of Medical Virology, Pathology Building, Prinshof Campus South, University of Pretoria, Private Bag X323, Gezina 0031, South Africa
Top